20210401 WGBS PicoMethyl-seq library prep for Thermal Transplant Trial 1
Project: Thermal Transplant Molecular
Goal
First trial of WGBS library prep using the Zymo Pico Methyl-seq library prep kit and this protocol.
Sample dilution and mix calculations can be found on this google sheet
Protocol
I am using a modified version of this protocol.
The only modification I made to this protocol was changing the input DNA from 1ng to 10ng. This worked in previous library preps performed by E. Strand and M. Schedl.
Samples and Indices
| Sample | Coral.ID | Life Stage | i5 index # | i7 index # |
|---|---|---|---|---|
| 18-358 | P-10-A | ADULT | 1 | 1 |
| 18-20 | R-7-B | ADULT | 2 | 2 |
| L-933 | P-10-A | LARVAE | 3 | 3 |
Qubit Results
To test DNA quantity: Qubit
I used the DNA broad range assay.
| Sample | DNA 1 (ng/ul) | DNA 2 (ng/ul) |
|---|---|---|
| Standard 1 | 184.99 | |
| Standard 2 | 19664.63 | |
| 18-358 (#1) | 21.6 | 21.0 |
| 18-20 (#2) | 12.2 | 11.8 |
| L-933 (#3) | 5.80 | 5.64 |
Tapestation Results
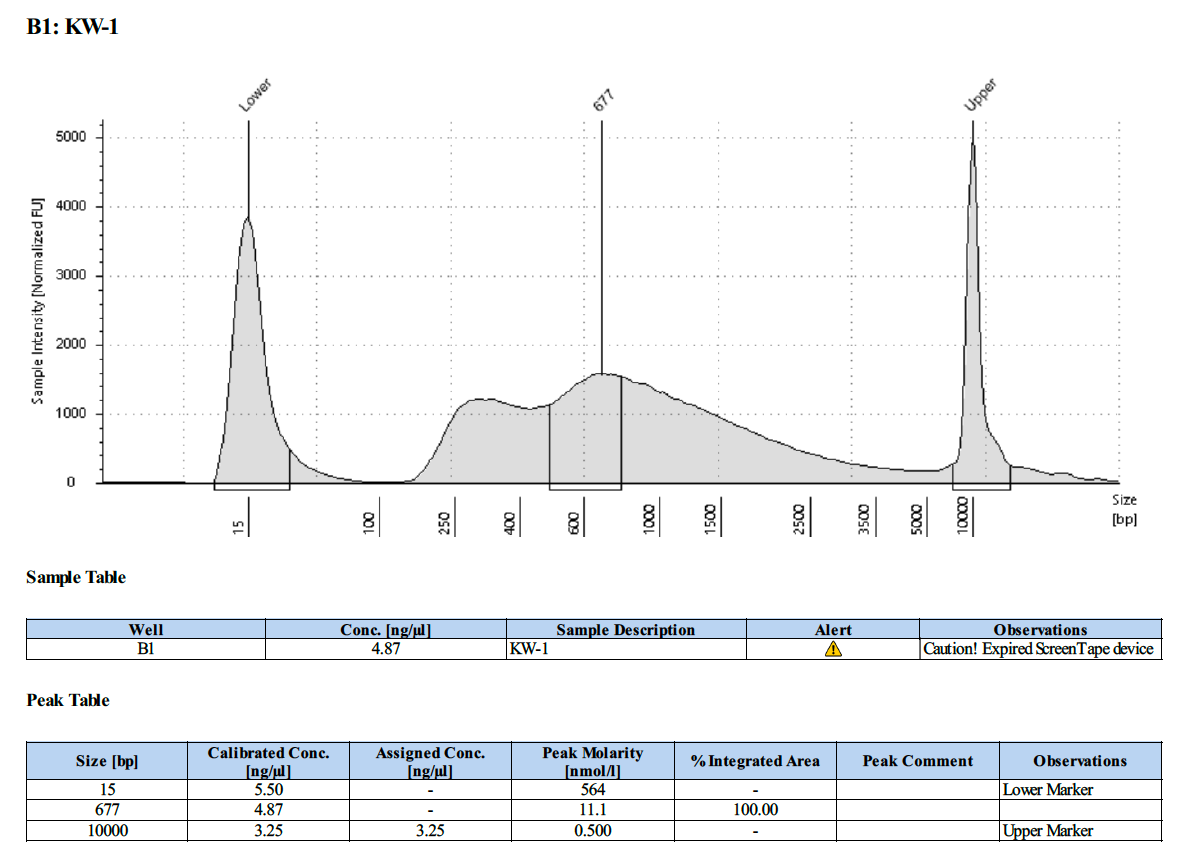
KW #1 (18-358):
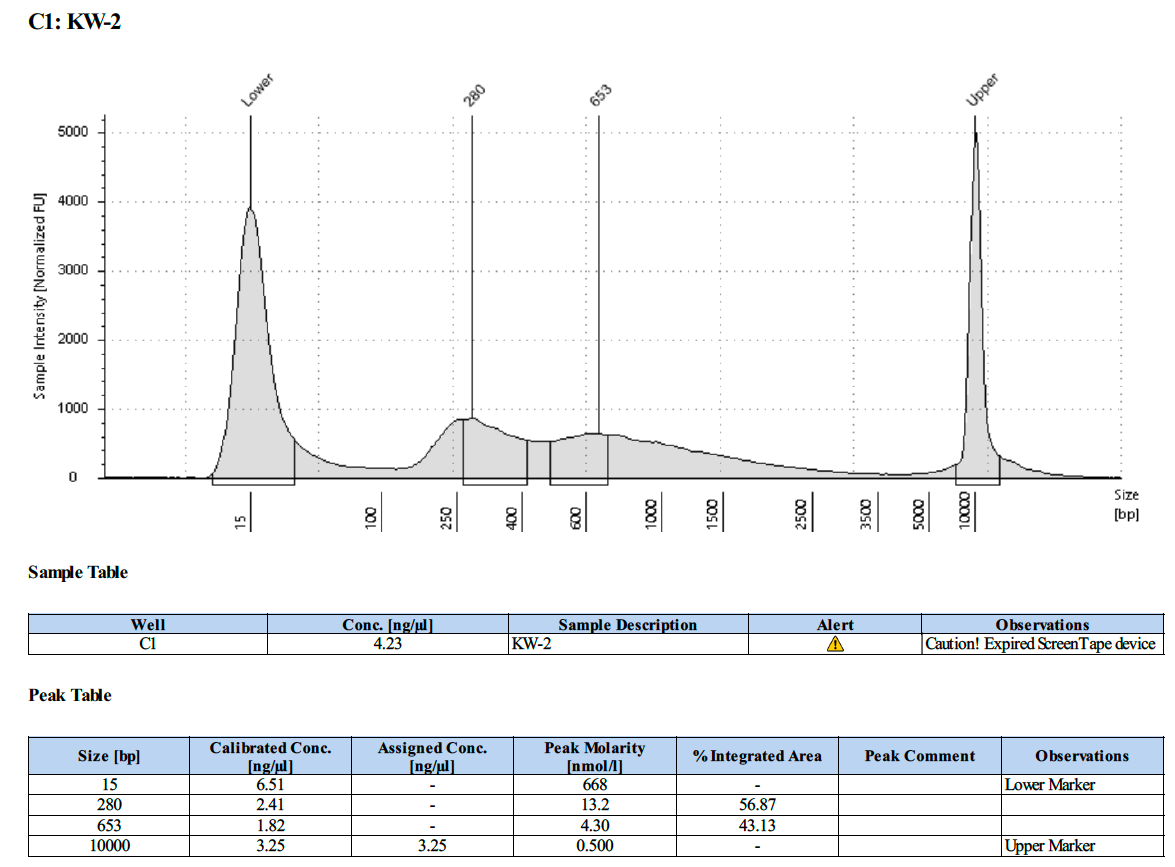
KW #2 (18-20):
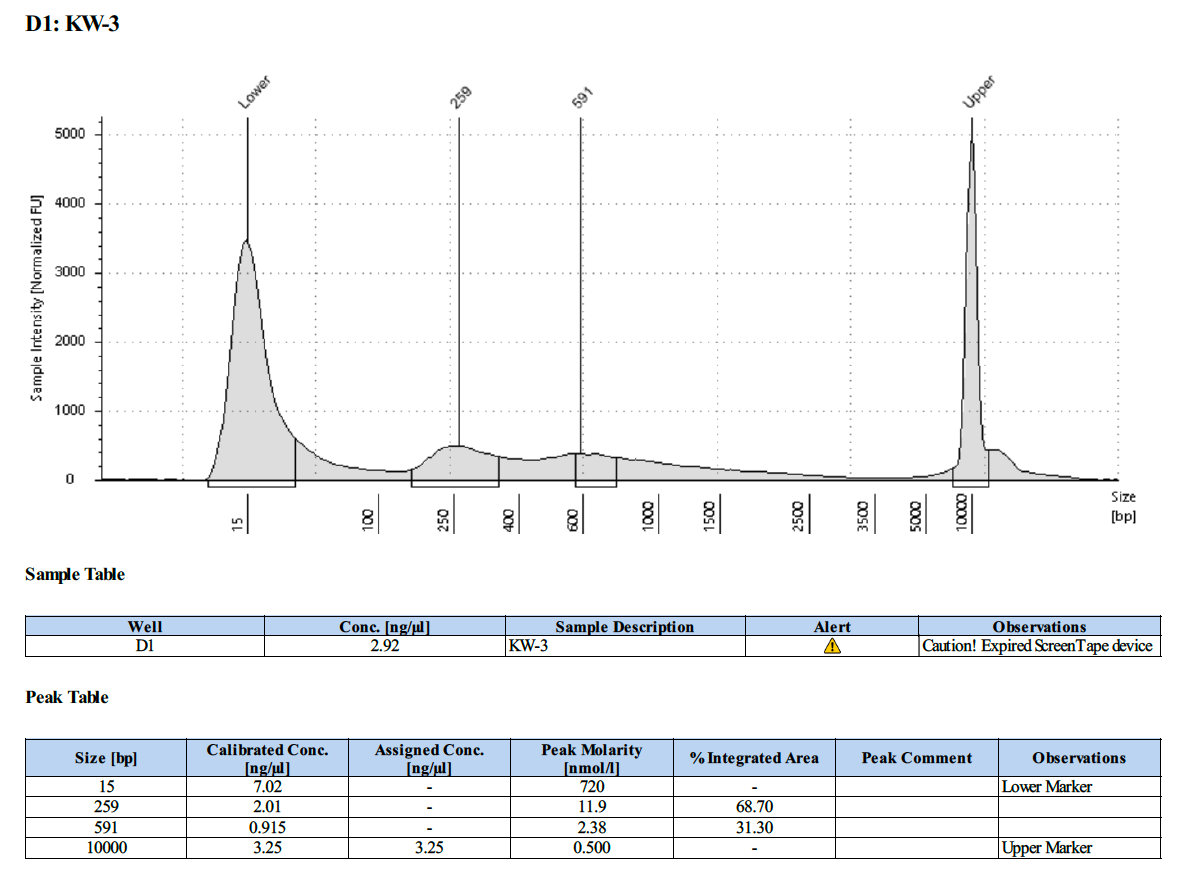
KW #3 (L-933):
Summary
Overall, this protocol worked, however there is PCR over-amplification. We will need to optimize the number number of PCR cycles to reduce this over-amplification.
Written on April 1, 2021
