20210622 Lipid Assay Trial
Goal
First attempt at the lipid assay using the unmodified protocol from Bove_Baumann_96well_Protocol.
The working Putnam Lab Lipid protocol is here
Overall Notes
- Pipetting the methanol and chloroform is very difficult! Work fast and angle the pipette (using filtered tips) to prevent dripping.
- Due to this error, I did not have enough to properly run the third triplicate of each standard.
- Next time, increase the volume of the standards so there is extra room for error.
- After the centrifugation, there will be three layers: the top layer (CHCl3), a middle layer that looks like mucus (aka interfacial fluff), and the bottom layer (lipid extract)
- This protocol produced EXACTLY 300uL of the lipid extract, so there is no room for error when removing the other two layers
- Fully remove the top layer (~675 uL) and leave the middle layer
- Gently move the middle layer to the side when pipetting out the bottom later.
- Tilting the vial to the side helps.
- This protocol produced EXACTLY 300uL of the lipid extract, so there is no room for error when removing the other two layers
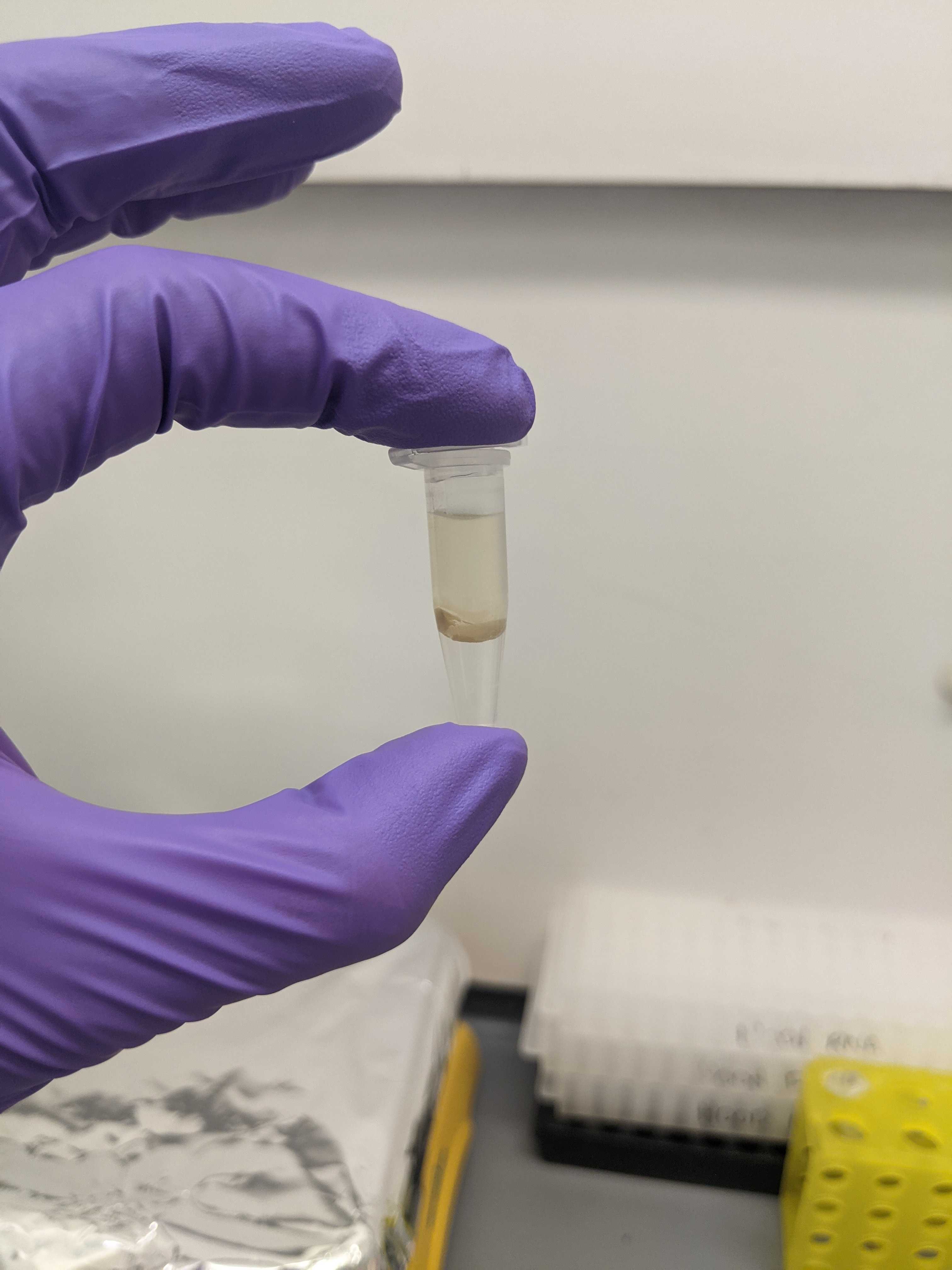
- After the H2SO4 addition and the 20 minute incubation at 90C, the liquid is very difficult to remove and I was not successfully able to retrieve 75uL out of each well. The bottom is very sticky and clogged the pipette. Tilting the plate helped to retrieve the liquid. Practice will definitely help.
- Fill the plate down the columns (i.e. A1-A12 then B1-B12) so you can use the multichannel
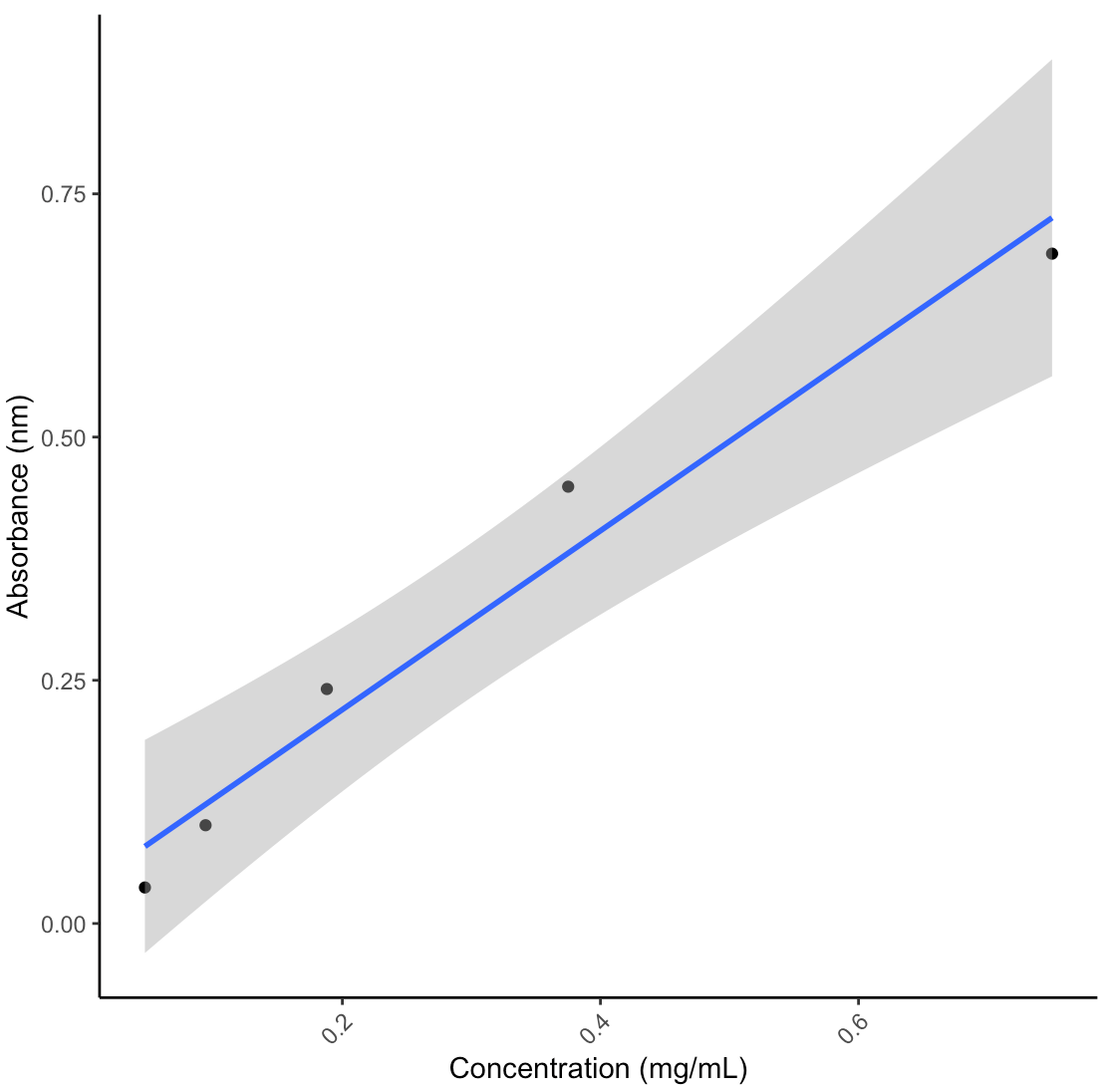
- With my homogenates and the 600uL input, my lipid concentrations were not high enough to detect a signal. The standard curve worked (after dropping the highest value due to pipette error), suggesting that the lipid assay worked but I will have to adjust the lipid extraction steps to accommodate my samples. The table below shows the calculated concentrations from the standard curve without any normalizations for dilutions, homogenate volume, or surface area yet.
- Next time, try increasing the input volume to 1mL of sample and keep the chloroform/methanol volumes the same
- We could also try vacuum centrifuging the samples to increase the concentration
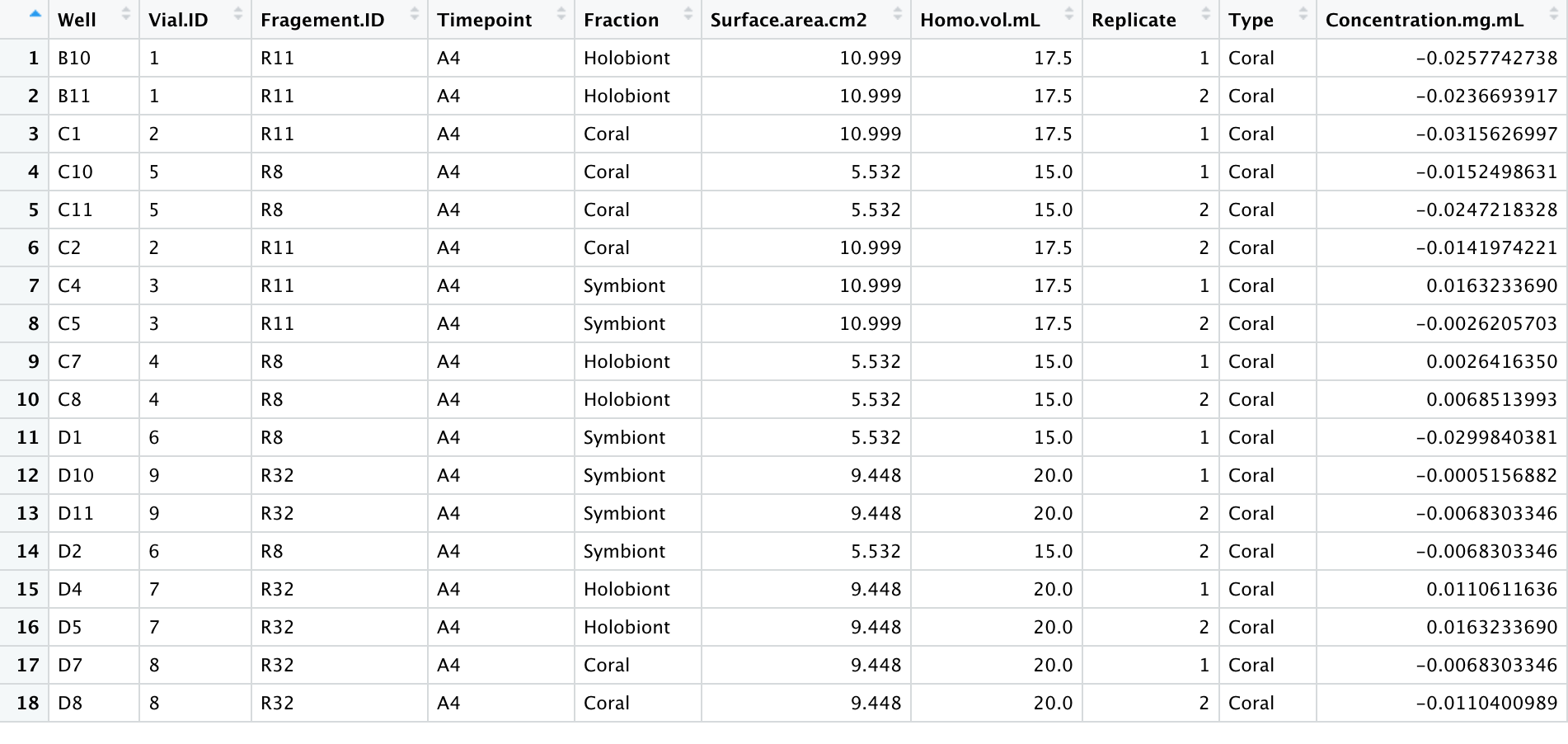
Samples
For this protocol, I used airbrushed homogenates from my Porites July Bleaching experiment using this protocol. These were extra coral homogenates that were frozen is 50mL falcon tubes.
Samples were taken out of the -80C freezer to thaw. Once thawed, 1 mL aliquots were put into the Holobiont and Symbiont tubes. The symbiont tubes were centrifuged for 3 minutes at 15000 rcf. The supernatant was transferred to the associated ‘coral’ tube and the pellet was resuspended in 1X PBS (phosphate buffer saline).
| Vial | Fragment.ID | Timepoint | Treatment | Fraction |
|---|---|---|---|---|
| 1 | R11 | A4 | Mortality | Holobiont |
| 2 | R11 | A4 | Mortality | Coral |
| 3 | R11 | A4 | Mortality | Symbiont |
| 4 | R8 | A4 | Bleached | Holobiont |
| 5 | R8 | A4 | Bleached | Coral |
| 6 | R8 | A4 | Bleached | Symbiont |
| 7 | R32 | A4 | Control | Holobiont |
| 8 | R32 | A4 | Control | Coral |
| 9 | R32 | A4 | Control | Symbiont |
Protocol used
Materials
Reagents:
Lipid extraction
- CH3OH (Methanol)
- CHCl3 (Chloroform)
- 0.05 M NaCl in water
Lipid Assay
- 17% Phosphoric acid (H3PO4)
- 0.2 mg/mL vanillin in 17% phosphoric acid
- Concentrated (18M) sulfuric acid (H2SO4)
- CH3OH (Methanol)
- CHCl3 (Chloroform)
- 1.5 mg/mL Corn Oil
Equipment:
- 96-well plates
- 1.5 mL tubes
- Vortex
- Plate shaker
- Centrifuge
- Hotplate
- Ice bucket/ice
- Fume hood
- Plate reader (can read absorbance at 540 nm)
Reagent and standard preparation
0.05 M NaCl:
- In a 50 mL labelled falcon tube,0.1461g NaCl in 50 mL DI water.
Stock 1.5 mg/mL Corn Oil
- In a 15 mL labelled falcon tube, add 245 μL of corn oil in 14.755 ml CHCl3
- Calculations:
- Density = 0.9188 g/ml (Noureddini et al., 1992)
- Total volume of ampule = 1 g* (1ml / 0.9188g) =1.08837614 ml
- Known concentration of ampule= 1000mg / 1.088ml = 918.8 mg/ml (same as known density)
- Therefore:
- (1.5 mg/mL * 15mL) / 918mg/mL = 0.0245 mL (or 245 μL) of 918mg/mL Corn oil standard concentrate
- Calculations:
Stock 0.2 mg/mL vanillin in 17% phosphoric acid (H3PO4):
- 20 mL of 17% H3PO4
- In a labelled 50 mL Falcon tube, add 4 mL 85% H3PO4 to 16 mL of DI water
- Stock vanillin solution
- In a labelled 50 mL Falcon tube, add 4 mg vanillin to 20 mL 17% H3PO4
Make subsets of the following reagents in labelled 50mL Falcon tubes daily
- CHCl3 (Chloroform)
- CH3OH (Methanol)
- Concentrated (18M) sulfuric acid (H2SO4)
Protocol
Note before starting: All portions of this protocol should be performed in a fume hood as much as possible with appropriate safety precautions, including gloves, lab coat, and closed toe shoes.
Lipid Extraction
- Pull samples from -80 freezer and allow to thaw
- Maximum of 24 samples per plate in triplicates
- Add 400 μL of CHCl3 and 200 μL of CH3OH in a 2:1 ratio to each labelled 1.5 mL sample tube
- Vortex sample and transfer 600 μL of coral tissue slurry sample to the corresponding 1.5 mL tube
- Vortex then shake on plate shaker for 20 minutes
- Prepare standards in the section below
- Add 160 μL of 0.05M NaCl
- CHCl3:CH3OH:NaCl is in a 2:1:0.8 ratio – keep this ratio
- Invert tubes gently two times and open and re-close lid
- Centrifuge at 3000 rpm for 5 minutes
- Remove CHCl3 (top) layer and dispose before taking 100 μL for the assay
- Do three times for 3 replicates of 100 μL
Lipid Standard creation
- Make a stock serial dilution in 7 1.5 mL tubes for each plate
- Add 300 μL of CHCl3 to standard tubes 2 through 7
- Add 600 μL of 1.5 mg/ml stock to standard tube 1
- Transfer 300 μL of tube 1 and place in tube 2. Pipette mix
- Pull 300 μL from tube 2 and place in tube 3. Pipette mix
- Repeat this process for tubes 3 through 6
- Do not add corn oil to tube 7! This is the blank
- Discard 300 μL from tube 6 so total volume equals 300 μL (optional)
| Standard | Concentration (mg/mL) |
|---|---|
| 1 | 1.5 |
| 2 | 0.75 |
| 3 | 0.375 |
| 4 | 0.188 |
| 5 | 0.094 |
| 6 | 0.047 |
| 7 (Blank) | 0 |
Lipid Assay
- In a 96-well plate, add 100 μL of sample or standard to each well in triplicates
- Add 50 μL of CH3OH to each well
- Evaporate solvent on a 90°C hotplate for 10 minutes
- Add 100 μL H2SO4 to each well
- Wells will change from clear to yellow colour
- Incubate on hotplate at 90°C for 20 minutes
- Cool the plate on ice for 2 minutes
- Transfer 75 μL of each sample or standard from the microplate to a new 96-well microplate
- Wells may contain sticky residue and bubbles, but you should be able to avoid that to pull the required 75 μL
- Cover the plate and read background absorbance at 540 nm using microplate reader
- This is a baseline measure that is used for correcting the final plate absorbance
- Add 34.5 μL of 0.2 mg/mL vanillin in 17% phosphoric acid to each well
- Incubate for 10 minutes
- Should change from yellow to pink color
- Cover the plate again and read absorbance at 540 nm using microplate reader
