20210826 Lipid Extraction Species Test
Goal
Attempting the lipid assay using the unmodified protocol from Bove_Baumann_96well_Protocol on multiple different coral species and host/symbiont fractions.
The working Putnam Lab Lipid protocol is here
Samples and Sample Preparation
For this protocol, I used airbrushed homogenates of Astrangia poculata (Proj; Astrangia Nutrition), Porites astreoides (Proj: Porites Nutrition 2019), Montipora capitata (Proj: Holobiont Integration), and Pocillopora acuta (Proj: Holobiont Integration). These were extra coral homogenates that were frozen is 50mL falcon tubes.
Samples were taken out of the -80C freezer to thaw. Once thawed, 1 mL aliquots were put into the Holobiont and Symbiont tubes. The symbiont tubes were centrifuged for 3 minutes at 15000 rcf. The supernatant was transferred to the associated ‘coral’ tube and the pellet was resuspended in 1X PBS (phosphate buffer saline).
| Vial | Fragment ID | Species | Fraction |
|---|---|---|---|
| 1 | 1246 | A. poculata (apo) | Coral |
| 2 | 1246 | A. poculata (apo) | Symbiont |
| 3 | 2012 | A. poculata (sym) | Coral |
| 4 | 2012 | A. poculata (sym) | Symbiont |
| 5 | P22-A2 | P. astreoides | Coral |
| 6 | P22-A2 | P. astreoides | Symbiont |
| 7 | M1822 | M. capitata | Coral |
| 8 | M1822 | M. capitata | Symbiont |
| 9 | P1170 | P. acuta | Coral |
| 10 | P1170 | P. acuta | Symbiont |
Results
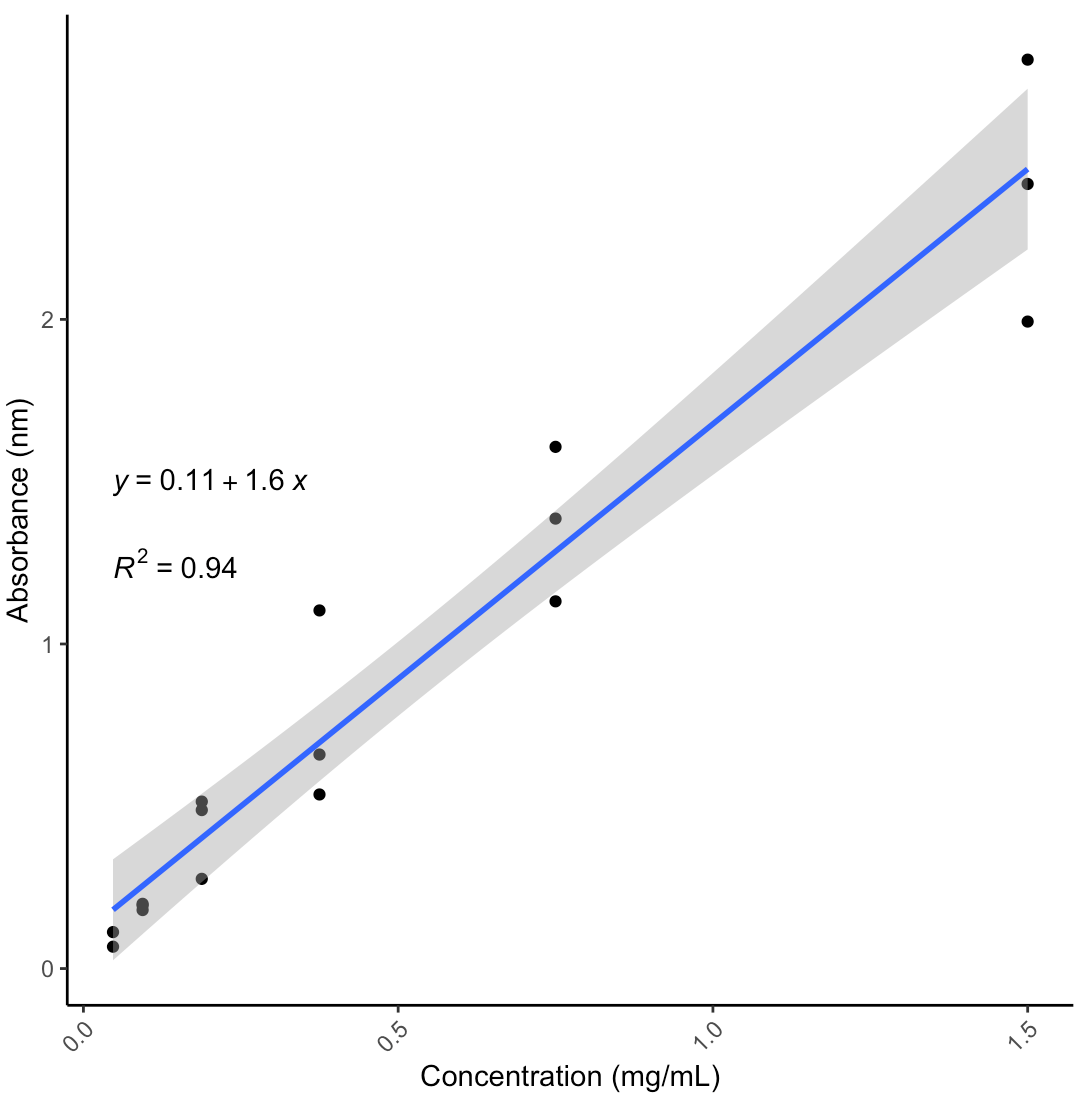
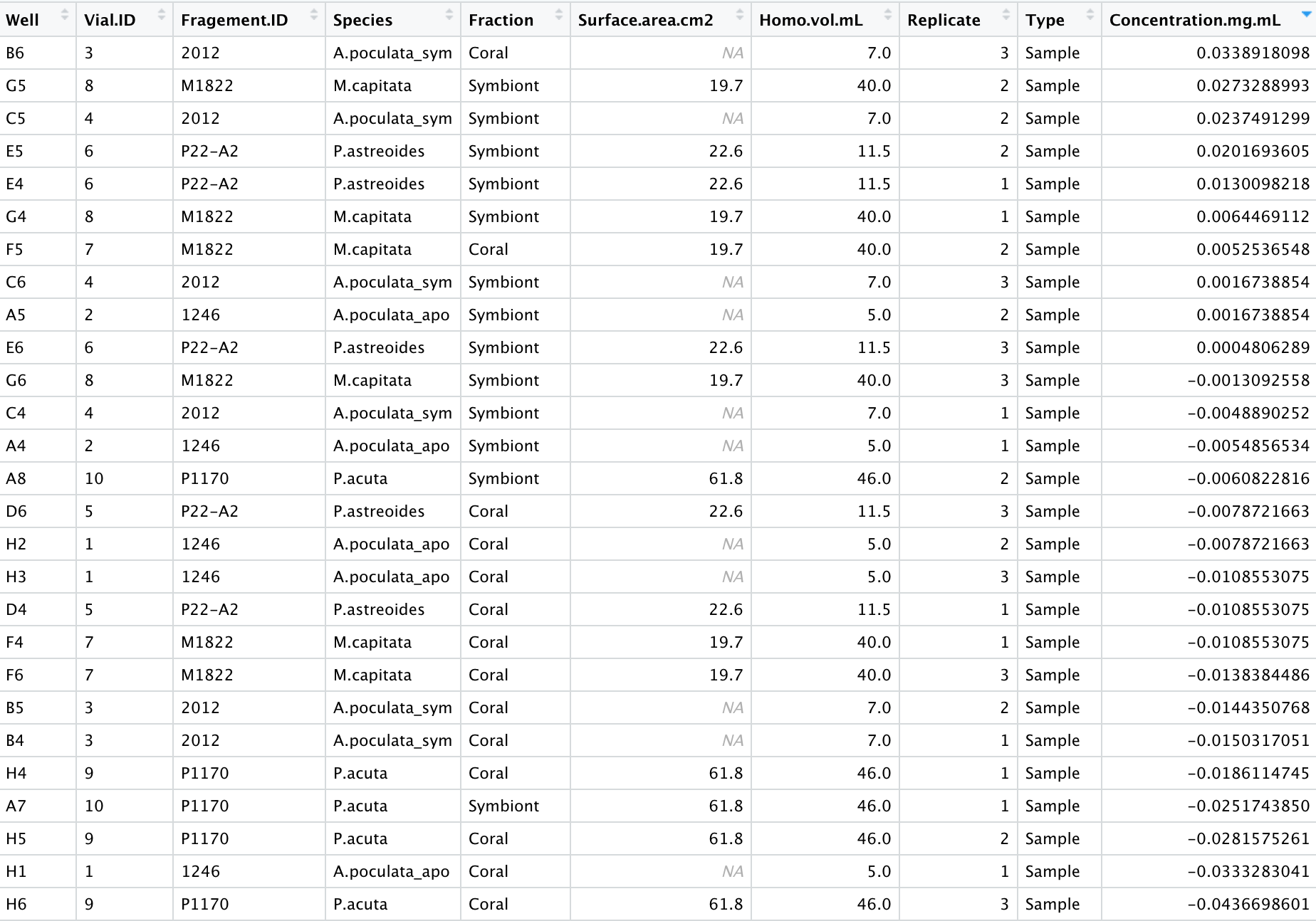
Notes
- After centrifugation in cholorform and methanol:
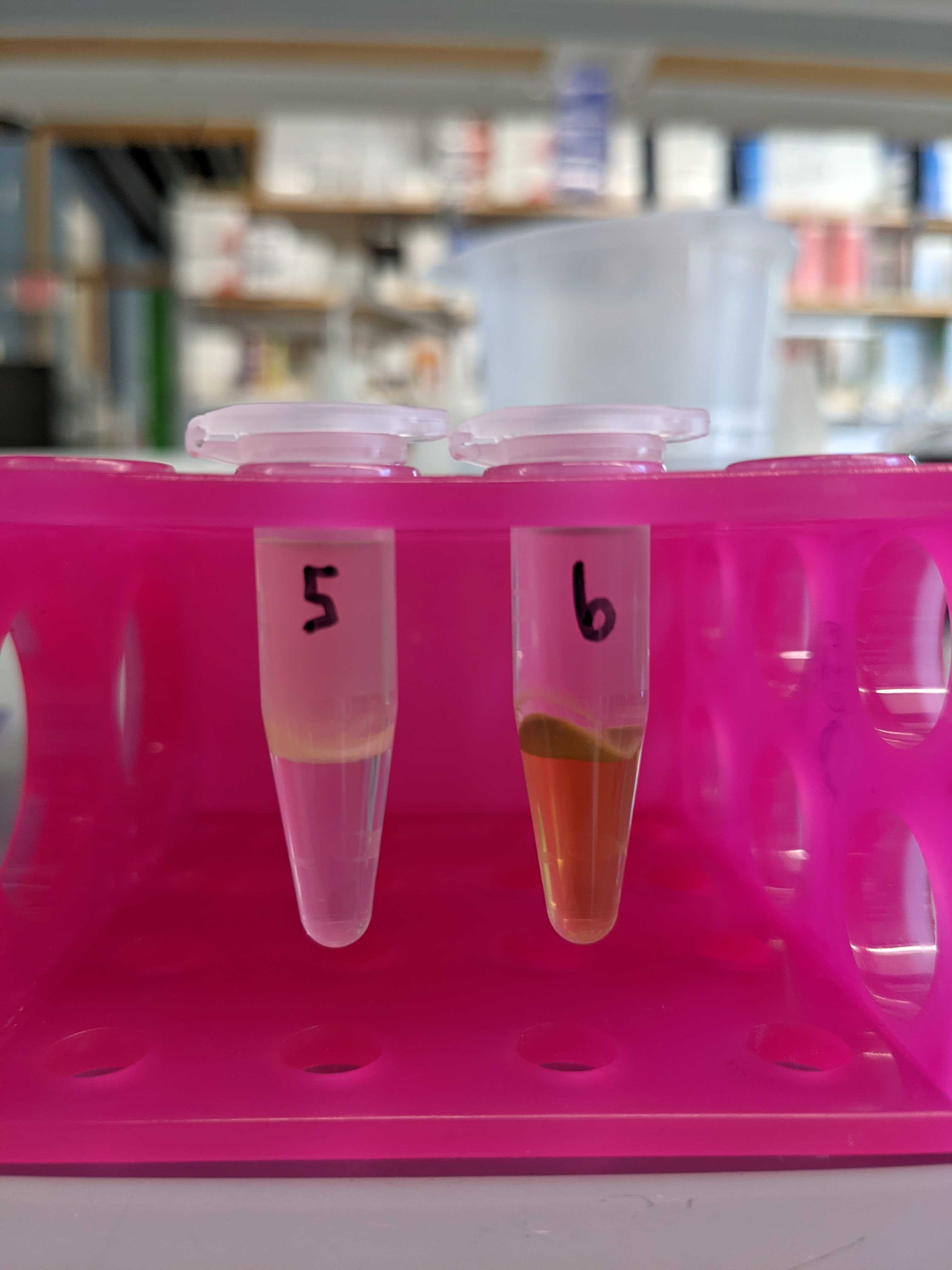
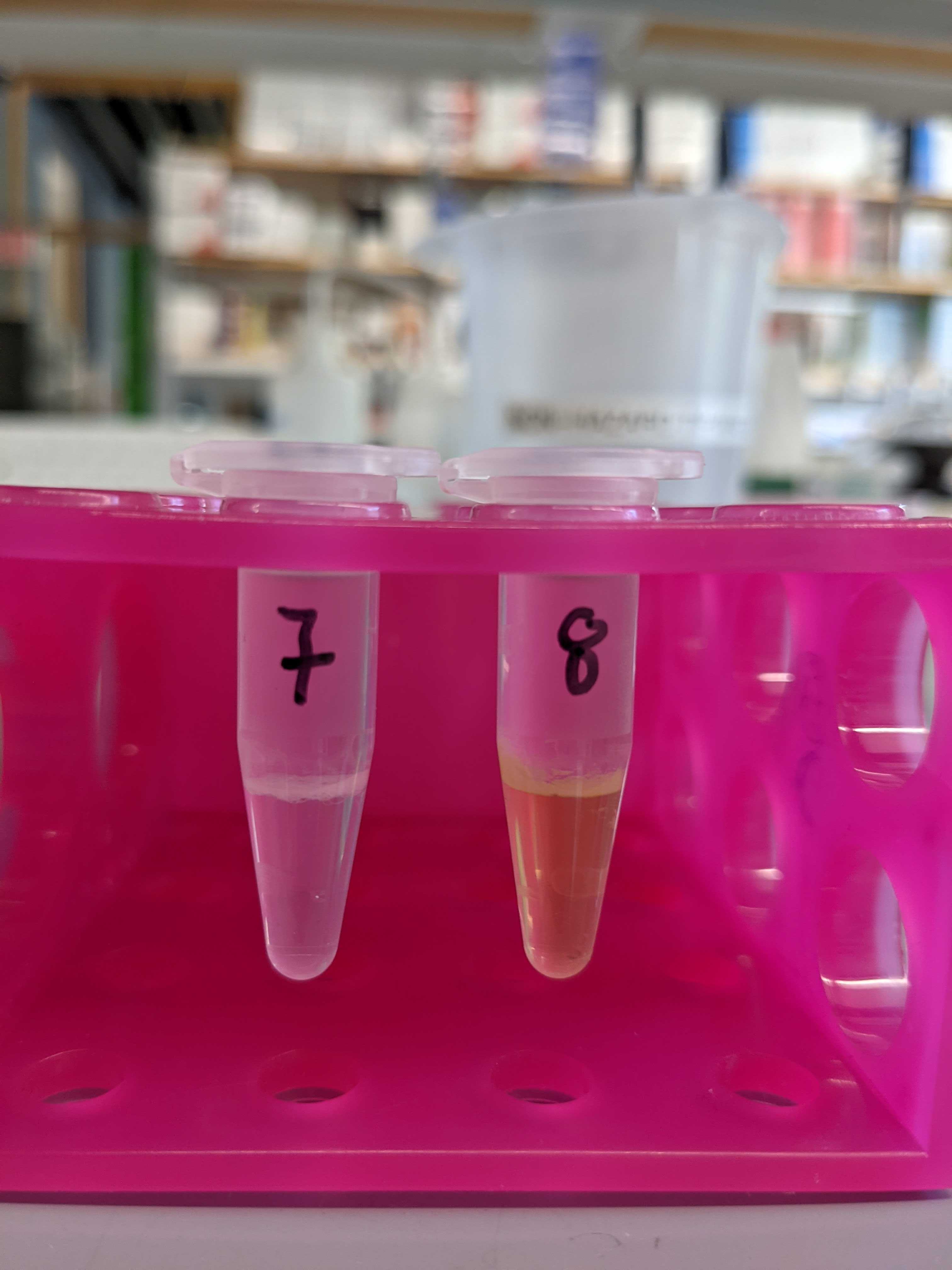
- P. astreoides coral fraction in the chloroform was still green
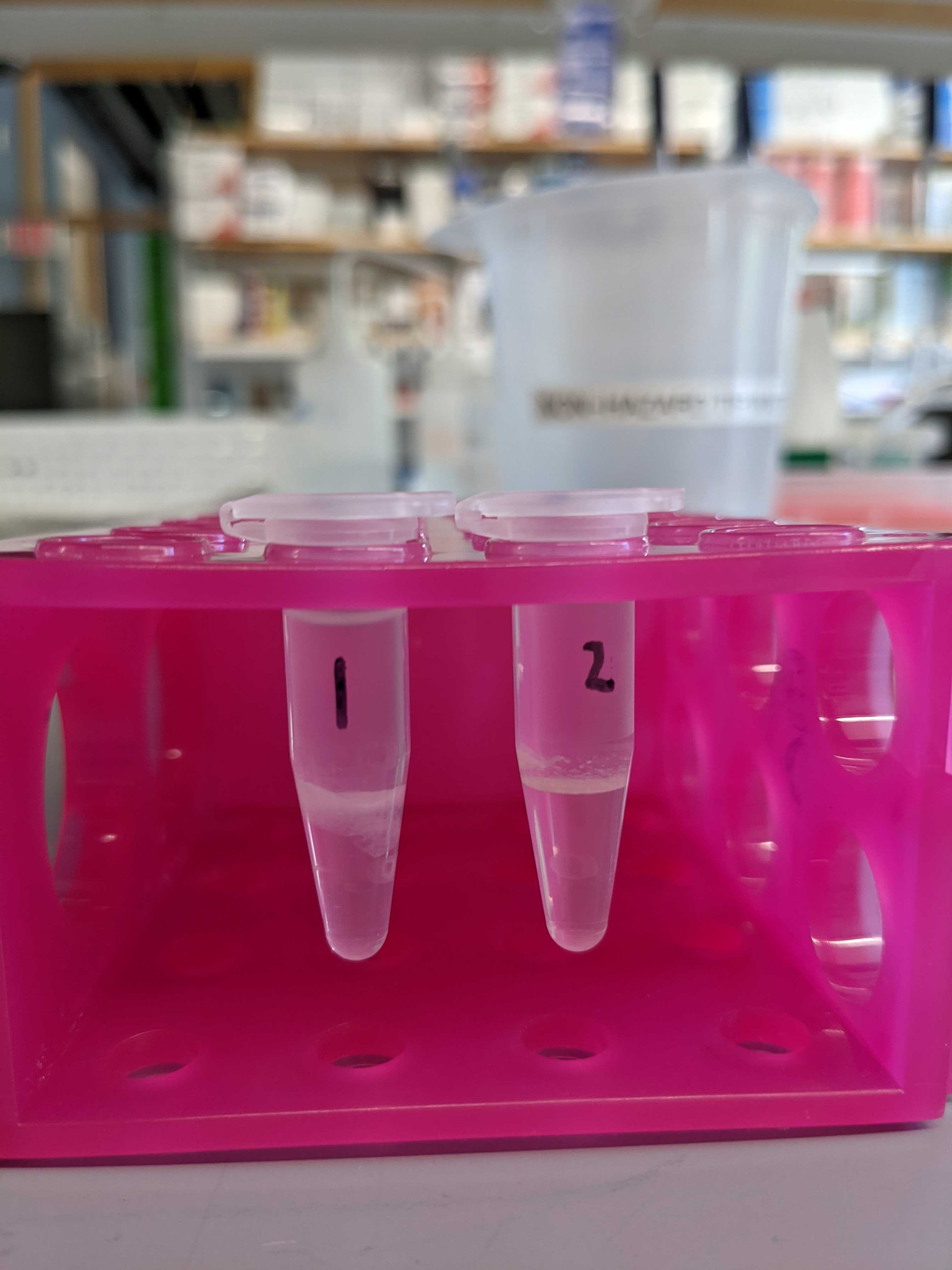
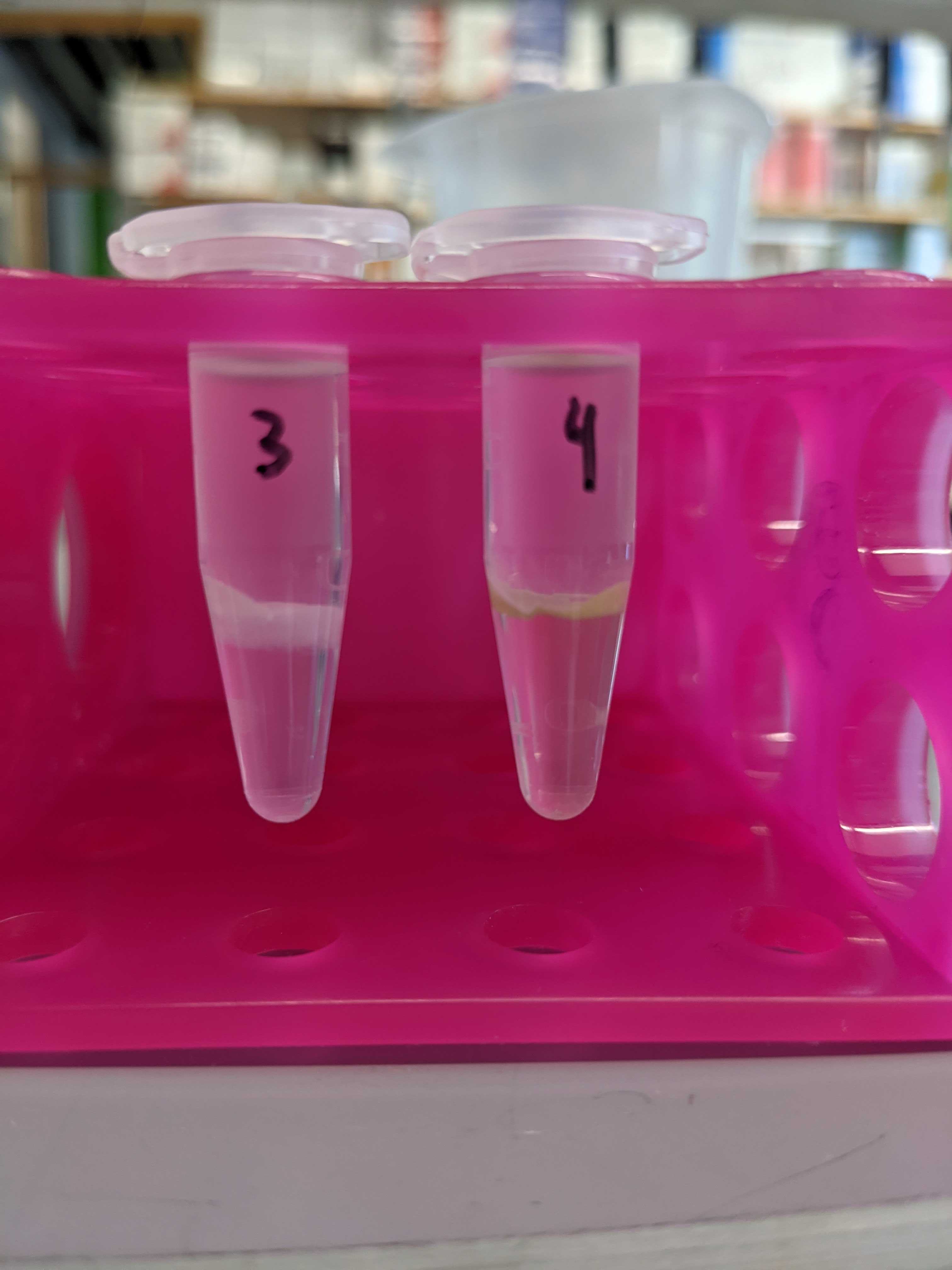
- Large “cellular debris” layer in some samples (see below)
- Samples 1 and 9 may have a bit of the cellular debris layer in the well due to pipette error (this actually resulted in positive readings I think)
- No samples in wells A6, A9, D5, F1 due to pipette error.
Below are photos of the samples after the shaker plate and centrifugation with chloroform and methanol.