20220319 PJB ITS2 re-seq
Goal: To re-extract DNA from symbiont pellets to increase chances of amplifying symbiont ITS2 region DNA rather than host mitochondria.
Project: Porites July Bleaching 2019
Samples
All samples were snap frozen, then airbrushed with 1X PBS (Phosphate Buffer Saline). 1 mL of the homogenate was aliquoted then re-frozen at -80 °C.
| Fragment_ID | Timepoint | Group | Day | Group_Day | Vial |
|---|---|---|---|---|---|
| R19 | A4 | Bleached | 52 | Bleached-52 | 132 |
| R29 | A4 | Bleached | 52 | Bleached-52 | 204 |
| R37 | A4 | Bleached | 52 | Bleached-52 | 168 |
| R8 | A4 | Bleached | 52 | Bleached-52 | 210 |
| R26 | A2 | Mortality | 37 | Mortality-37 | 138 |
| R28 | A2 | Mortality | 37 | Mortality-37 | 72 |
| R36 | A2 | Mortality | 37 | Mortality-37 | 12 |
| R28 | A4 | Mortality | 52 | Mortality-52 | 144 |
| R35 | A4 | Mortality | 52 | Mortality-52 | 84 |
| R36 | A4 | Mortality | 52 | Mortality-52 | 120 |
| R7 | A4 | Control | 52 | Control-52 | 78 |
DNA Extraction (performed on 20220319)
Sample Preparation and Digestion
Following the Biological fluids and cell protocol from the Zymo Quick DNA extraction kit:
- Take samples out from -80 °C
- Centrifuge samples for 3 minutes at 9,000 rcf and remove supernatant
- Resuspend the pellet with 200uL of 1X PBS
- Immediately added 200uL Biofluid Cell Buffer (red) and 20 μl of Proteinase K to each sample
- Vortex and spin down
- Incubate for 30 minutes at 55 °C on 1100 rpm
- Centrifuge at 8,000 rcf for 30 seconds to remove debris
- Transfer 350 μl of the supernatant to a new, labelled 1.5 ml centrifuge tube
DNA Extraction
- Add equal volume (350 uL) of Genomic Binding Buffer to each tube
- Finger flick to mix
- Add 700 uL to the spin column
- Centrifuge at 12,000 g (rcf) for 1 minute
- Add 400 uL pre-wash buffer to each spin column
- Centrifuge at 12,000 g (rcf) for 1 minute
- Remove and discard flow through
- Add 700 uL wash buffer to each spin column
- Centrifuge at 12,000 g (rcf) for 1 minute
- Remove and discard flow through
- Add 200 uL wash buffer to each spin column
- Centrifuge at 12,000 g (rcf) for 2 minute
- Transfer spin columns to new 1.5 mL centrifuge tubes
- Add 50 uL of warmed 10mM DNA Elution buffer directly to the filter in the spin column
- Incubate at room temperature for 15 minutes
- Centrifuge at 12,000 rcf for 1 minute
- Add another 50 uL of warmed 10mM DNA Elution buffer directly to the filter in the spin column
- Incubate at room temperature for 5 minutes
- Centrifuge at 12,000 rcf for 1 minute
- Label final tubes
- Store labelled samples in -20 °C
Quantify Results
Qubit
DNA: Broad Range
| Sample | DNA #1 (ng/uL) | DNA #2 (ng/uL) |
|---|---|---|
| ST 1 | 193.08 | |
| ST2 | 19841.21 | |
| 12 | 7.88 | 8.2 |
| 72 | 21.6 | 22.0 |
| 78 | 45.2 | 45.8 |
| 84 | 21.2 | 21.2 |
| 120 | 10.0 | 10.5 |
| 132 | 39.2 | 39.8 |
| 138 | 21.4 | 21.8 |
| 144 | 22.4 | 22.8 |
| 168 | 13.1 | 13.2 |
| 204 | 19.8 | 20.2 |
| 210 | 12.8 | 12.9 |
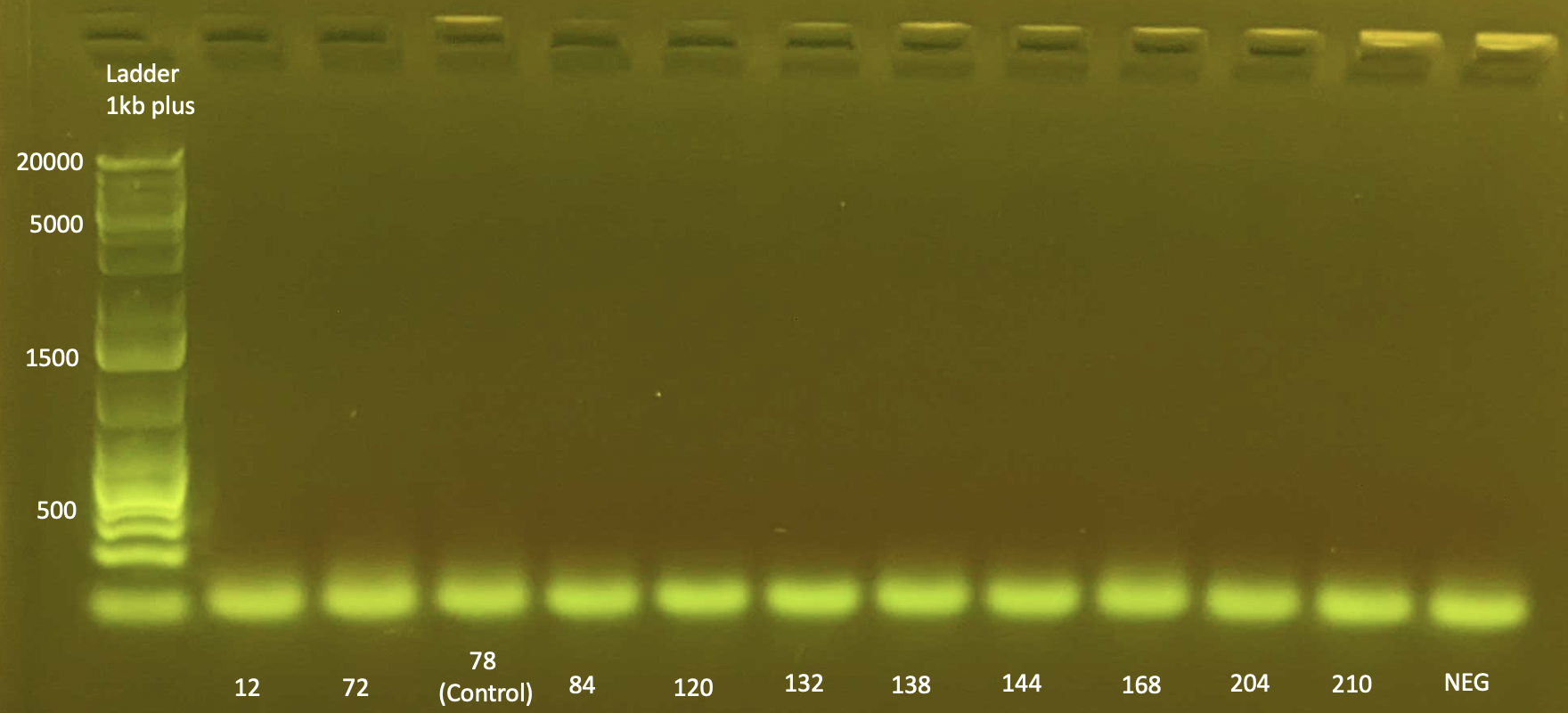
Gel Electrophoresis
To test DNA quality: Gel Electrophoresis
- Today I did a small gel of the above protocol and ran it at 100V.
ITS2 (performed on 20220321)
Dilution Planning
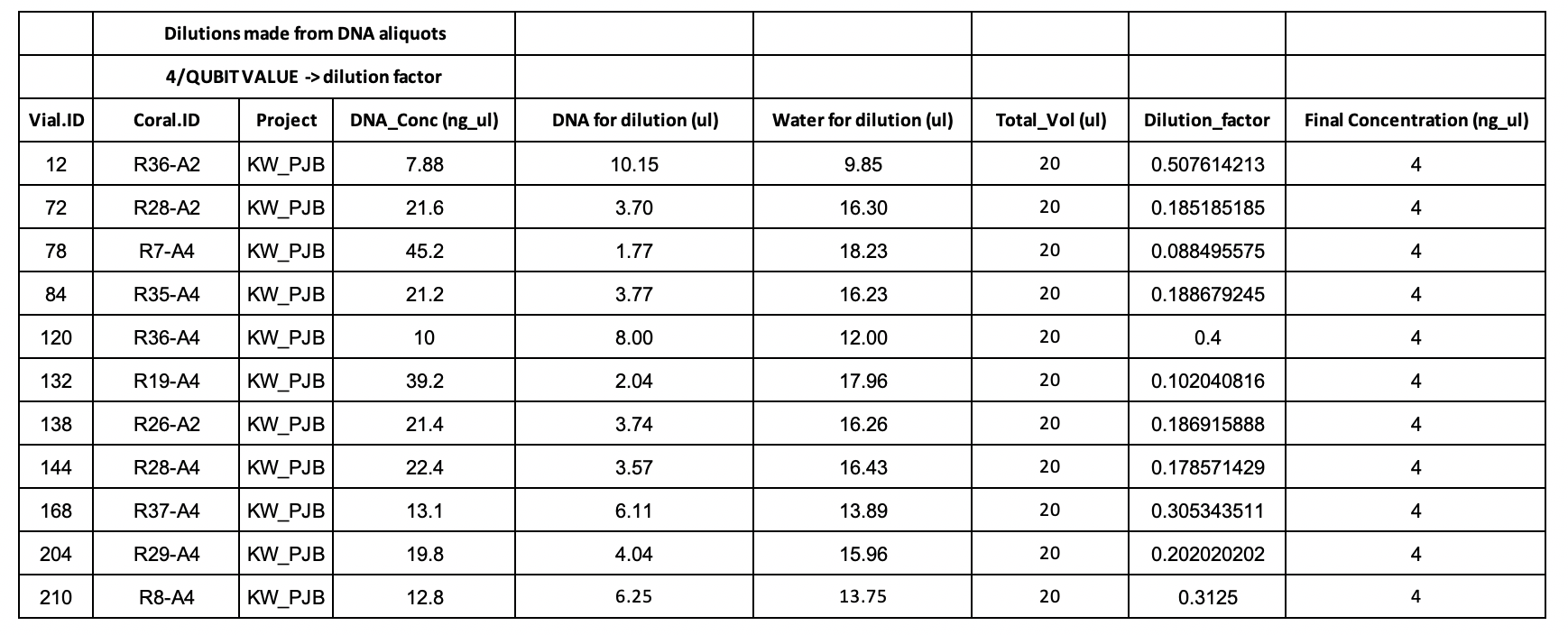
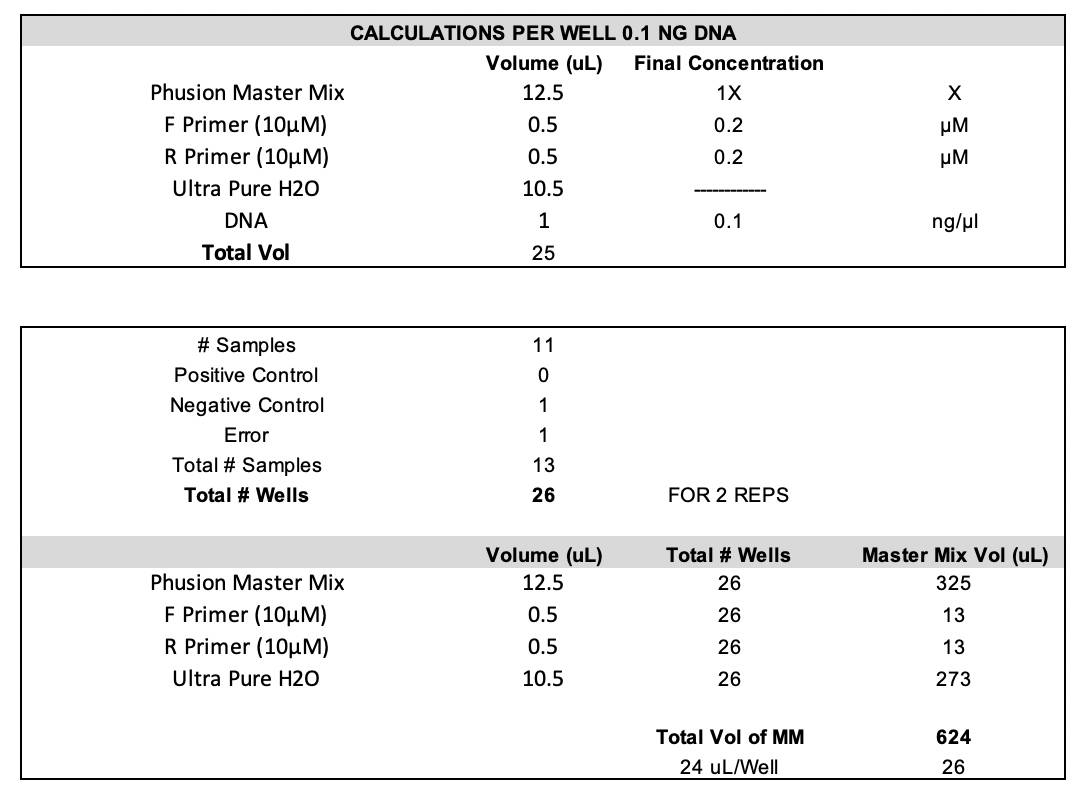
Protocol and Master Mix Planning
I am follow the Putnam Lab’s original ITS2 protocol.
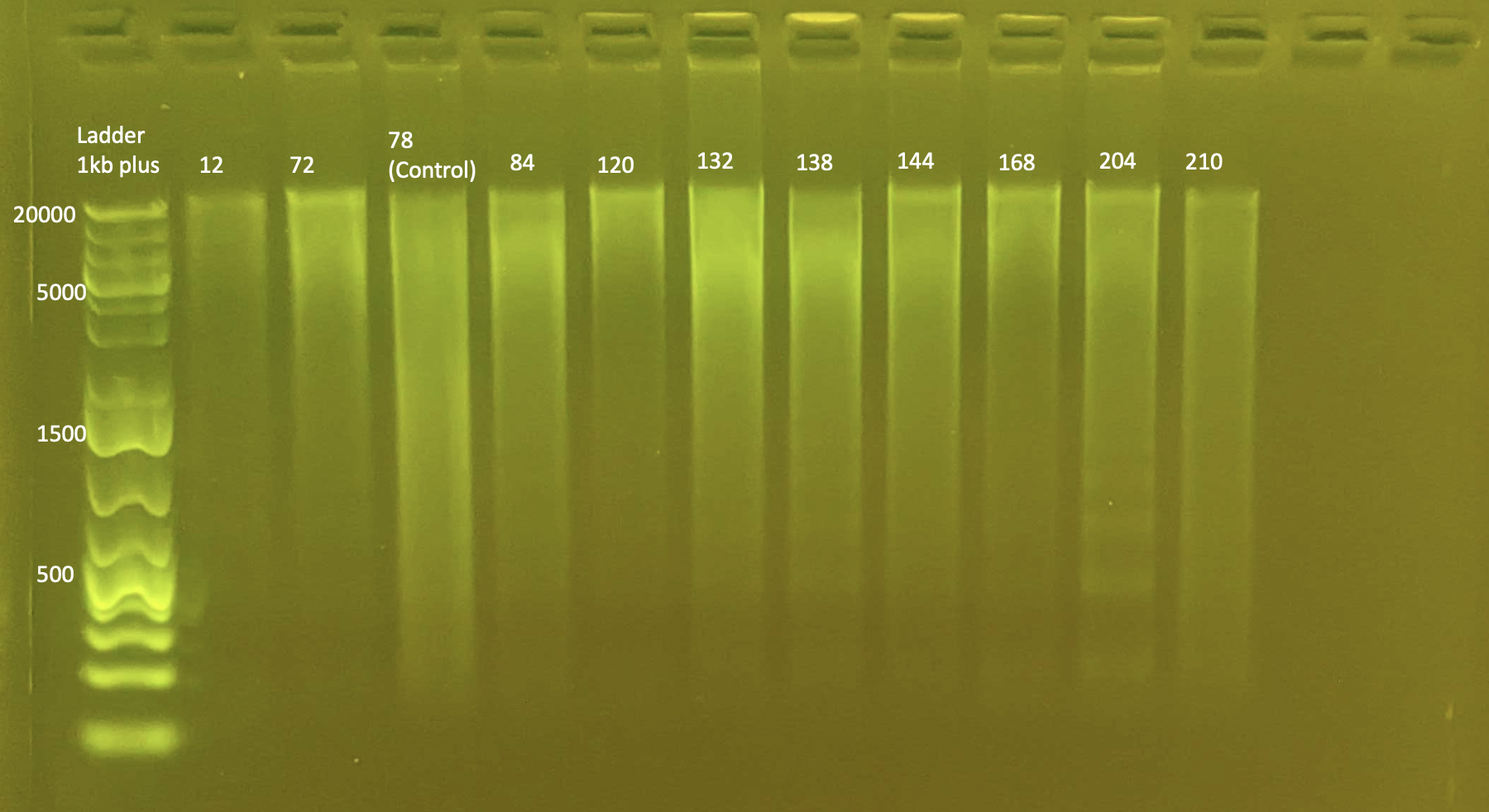
Gel Electrophoresis
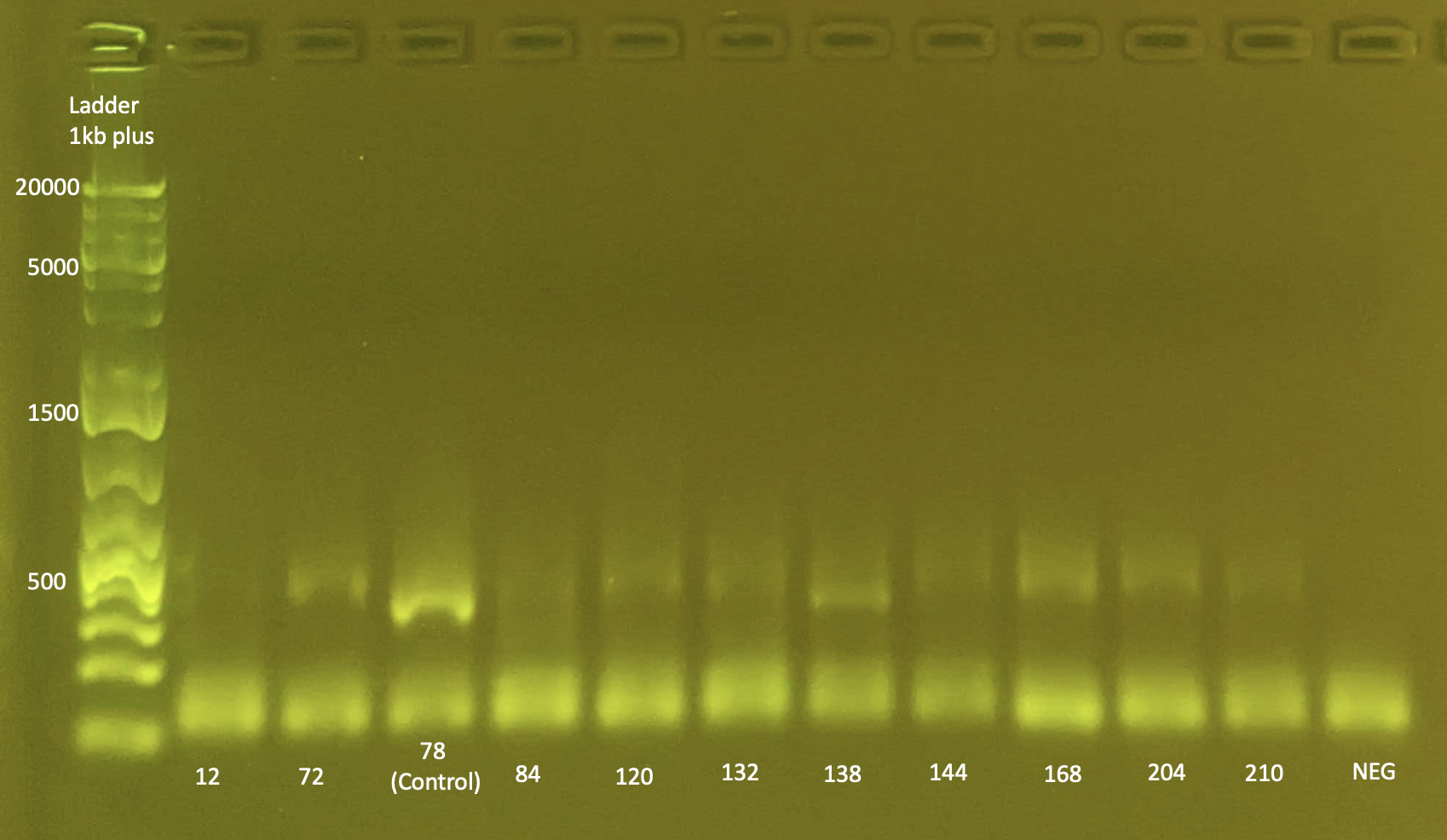
To test ITS2 amplification: Gel Electrophoresis
- Today I did a small gel of the above protocol and ran it at 100V.
ITS2 (performed on 20220420)
I ran the same protocol as above except with the following thermocyler settings:
| Cycles | Time | Temp | |
|---|---|---|---|
| 1 | 3 min | 95° | |
| 35 | 30 sec | 95° | |
| 30 sec | 71° | ||
| 30 sec | 72° | ||
| 1 | 2 min | 72° | |
| 1 | ∞ min | 4° | s |
I decided to change the annealing temperature to 71° because this is the calculated annealing temperature for the primer with the Illumina adapters.
Gel Electrophoresis
To test ITS2 amplification: Gel Electrophoresis
- Today I did a small gel of the above protocol and ran it at 75V.