DNA-RNA Extractions Porites astreoides homogenates, larvae, and adult fragments
Goal
To test the protocol for DNA/RNA extractions for airbrushed homogenates, larvae, and adult fragments of Porites astreoides for the Thermal Transplant 2017/2018 experiment.
Samples
I used 3 adult P. astreoides samples that were snap frozen, then airbrushed with filtered seawater. 0.5 mL of the homogenate was aliquoted then re-frozen at -80 °C. I also used 3 larval samples from 2018. 20 larvae were samples per vial. Additionally, I used 3 fragmented adult coral chips that were snap frozen, chipped, and re-frozen with 500uL of DNA/RNA shield and beads.
Sample Numbers
- 17-215
- Adult, R4-T2 (2017: Rim + Ambient Treatment)
- Homogenate
- 17-233
- Adult, R11-T2 (2017: Rim + High Treatment)
- Homogenate
- 17-521
- Adult, P19-T2 (2017: Patch + High Treatment)
- Homogenate
- 17-530
- Adult, P6-T2 (2017: Patch + Ambient Treatment)
- Homogenate
- L-534
- Larvae, R11-A (2018: Rim + High Treatment + Patch Transplant)
- L-939
- Larvae, P12-A (2018: Patch + High Treatment + Patch Transplant)
- L-1026
- Larvae, P16-A (2018: Patch + High Treatment + Patch Transplant)
- 18-31-C
- Adult, P9-A (2018: Patch + Ambient Treatment + Patch Transplant)
- Fragment
- 18-43-C
- R15-B (2018: Rim + High Treatment + Rim Transplant)
- Fragment
- 18-57-C
- P19-A (2018: Patch + High Treatment + Patch Transplant)
- Fragment
Protocol description
I used a modified version of this protocolfor homogenates, this protocol for larvae, and this protocol for fragments.
Homogenate preparation
- Immediately after taking sample out of the -80°C freezer, add 500μL of DNA/RNA shield
- Once fully thawed, add 100 μL of Protenase K digestion buffer (Derived from volume of homogenate and 300/30 ratio from Zymo Protocol) and 50 μL of Protenase K (Derived from volume of homogenate and 300/15 ratio from Zymo Protocol)
- Vortex and spin down.
- Incubate on Thermomixer at 56°C at 1500 rpm for 30 minutes
- Spin down for 30 seconds to pellet any debris
- Remove 1000 μL of the supernatant and transfer in 5 mL tubes. Avoid any debris.
Larvae preparation
- Immediately after taking sample out of the -80°C freezer, add 1000μL of DNA/RNA shield
- Once fully thawed, add 100 μL of Protenase K digestion buffer (Derived from volume of homogenate and 300/30 ratio from Zymo Protocol) and 50 μL of Protenase K (Derived from volume of homogenate and 300/15 ratio from Zymo Protocol)
- Vortex and spin down.
- Incubate on Thermomixer at 56°C at 1500 rpm for 90 minutes
- Spin down for 30 seconds to pellet any debris
- Remove 1000 μL of the supernatant and transfer in 5 mL tubes. Avoid any debris.
Fragment preparation
- Take samples out from -80 °C.
- Add 500 μl of RNA/DNA shield to make the total volume 1 mL
- These samples already have beads in them.
- Vortex for 2 minutes. Leave the settings on and on max power.
- Remove 700 μl of the supernatant from the bead tube and place in a new 1.5 microcentrifuge tube labeled on the side with the extraction sample number and today’s date. Label the cap of the microcentrifuge tube with the sample number.
- Add 70 μl of Proteinase K digestion buffer (10:1 ratio of sample:digestion buffer), and 35 μl of Proteinase K (2:1 ratio of digestion buffer:Proteinase K) to the 5 mL centrifuge tube.
- Incubate on Thermomixer at 56°C at 1500 rpm for 30 minutes
- Spin down for 30 seconds to pellet any debris
- Transfer 700 μl of the supernatant to a new, labelled 5 ml centrifuge tube.
- Vortex and spin down all tubes.
Zymo Duet RNA DNA Extractions
Modified from the Zymo protocol.
DNA Extraction
- Add equal volume of DNA lysis buffer as sample volume to the 5 mL tube with sample
- Vortex to mix
- Transfer 700 μL into DNA filter column (yellow)
- Centrifuge at 16,000 rcf for 30 seconds
- Remove flow through liquid and transfer into a new 5 mL tube labeled for RNA
- Repeat steps 2 until all liquid is gone
- Add 400 μL of DNA/RNA prep buffer to column
- Centrifuge at 16,000 rcf for 30 seconds
- Remove the flow through and transfer to waste
- Add 700 μL of DNA/RNA wash buffer
- Centrifuge at 16,000 rcf for 30 seconds
- Remove the flow through and transfer to waste
- Add 400 μL of DNA/RNA wash buffer
- Centrifuge at 16,000 rcf for 2 minutes
- Remove the flow through and transfer to waste
- Removed column and place into a new sterile 1.5 mL tube
- Add 50 μL of 10 mM Tris HCL (warmed to 55°C) directly to filter
- Incubate at room temperature for 15 minutes
- Centrifuge at 16,000 rcf for 30 seconds
- Keep flow through in tube
- Add another 50 μL of 10 mM Tris HCL (warmed to 55°C) directly to filter
- Incubate at room temperature for 5 minutes
- Centrifuge at 16,000 rcf for 30 seconds
- Discard column and keep flow through in tube
- Aliquot 10 μL into PCR tubes for QC.
- Store remaining aliquot at -20°C
RNA Extraction
- Add equal volume of 100% Ethanol as sample volume to the 5 mL tube with sample
- Vortex to mix
- Transfer 700 μL into RNA filter column (green)
- Centrifuge at 16,000 rcf for 30 seconds
- Remove flow through liquid and transfer into a new 5 mL tube labeled for Protein
- Repeat steps 12 until all liquid is gone
- Add 400 μL of DNA/RNA wash buffer to column
- Centrifuge at 16,000 rcf for 30 seconds
- Remove the flow through and transfer to waste
-
Make DNase I master mix: [75μL DNA digestion buffer and 5μL DNase] X n (sample #)
- Add 80μL of DNase I master mix directly to the filter of each column. Incubate at room temperature for ~15 minutes
- Add 400 μL of DNA/RNA prep buffer to column
- Centrifuge at 16,000 rcf for 30 seconds
- Remove the flow through and transfer to waste
- Add 700 μL of DNA/RNA wash buffer
- Centrifuge at 16,000 rcf for 30 seconds
- Remove the flow through and transfer to waste
- Add 400 μL of DNA/RNA wash buffer
- Centrifuge at 16,000 rcf for 2 minutes
- Remove the flow through and transfer to waste
- Removed column and place into a new sterile 1.5 mL tube
- Add 50 μL of RNAase-free water (warmed to 55°C) directly to filter
- Incubate at room temperature for 5 minutes
- Centrifuge at 16,000 rcf for 30 seconds
- Keep flow through in tube
- Made the gel during this incubation
- Add another 50 μL of RNAase-free water (warmed to 55°C) directly to filter
- Incubate at room temperature for 5 minutes
- Centrifuge at 16,000 rcf for 30 seconds
- Discard column and keep flow through in tube
- Took out qubit standards
- Aliquot 5 μL in PCR strip tubes for QC.
- Store remaining aliquot at -80 °C
Clean-up
- Place tissue and liquid in the waste container labeled Zymo extraction waste.
- Wipe down RNA free area with RNase away and kimwipes.
- Throw away all tips and restock tip boxes if necessary.
Testing Quantity and Quality
Quantify Results
Qubit
To test RNA and DNA quantity: Qubit
10 samples + 2 standards + 1 error = 13
DNA Broad Range
199 µl Buffer x 13 = 2587 µl Buffer 1 µl Reagent x 13 = 13 µl Reagent
RNA Broad Range
199 µl Buffer x 13 = 2587 µl Buffer 1 µl Reagent x 13 = 13 µl Reagent
| Sample Type | Sample | DNA (ng/uL) | RNA (ng/uL) |
|---|---|---|---|
| Standard 1 | 188.26 | 393.91 | |
| Standard 2 | 19093.72 | 8966.36 | |
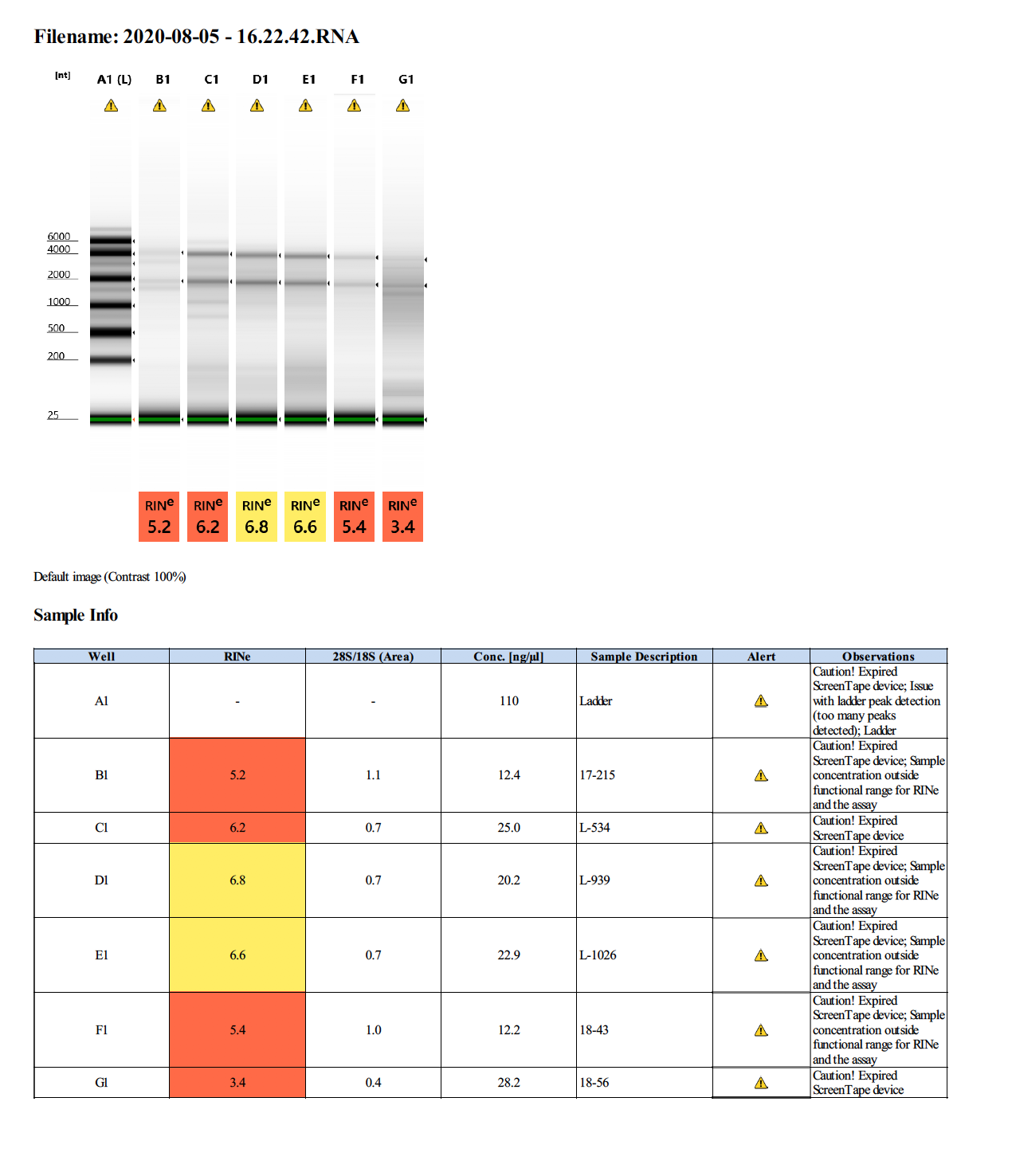
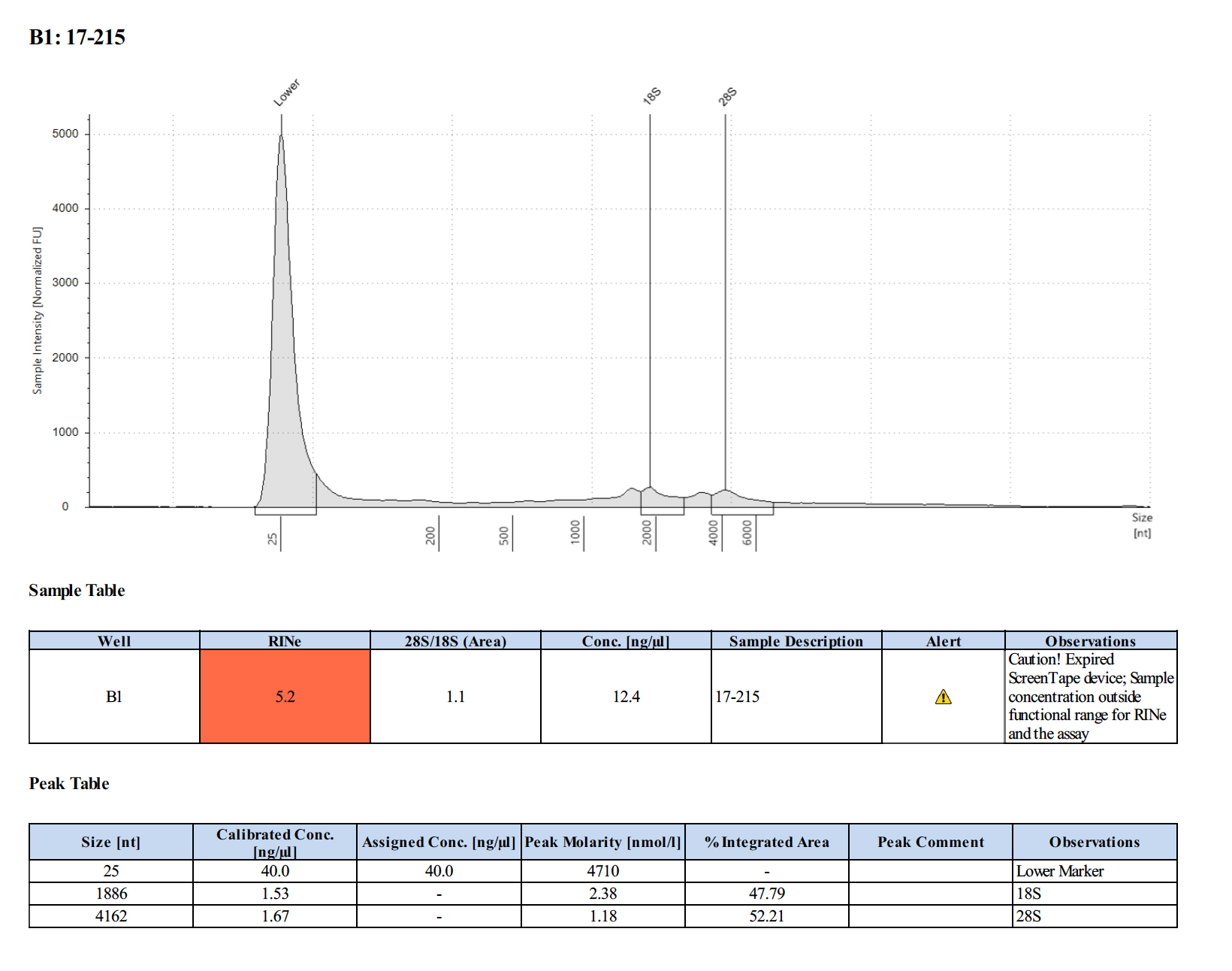
| Homogenate | 17-215 | 27.4 | 15.8 |
| Homogenate | 17-233 | 5.72 | LOW |
| Homogenate | 17-521 | 11.6 | LOW |
| Homogenate | 17-330 | 28.0 | LOW |
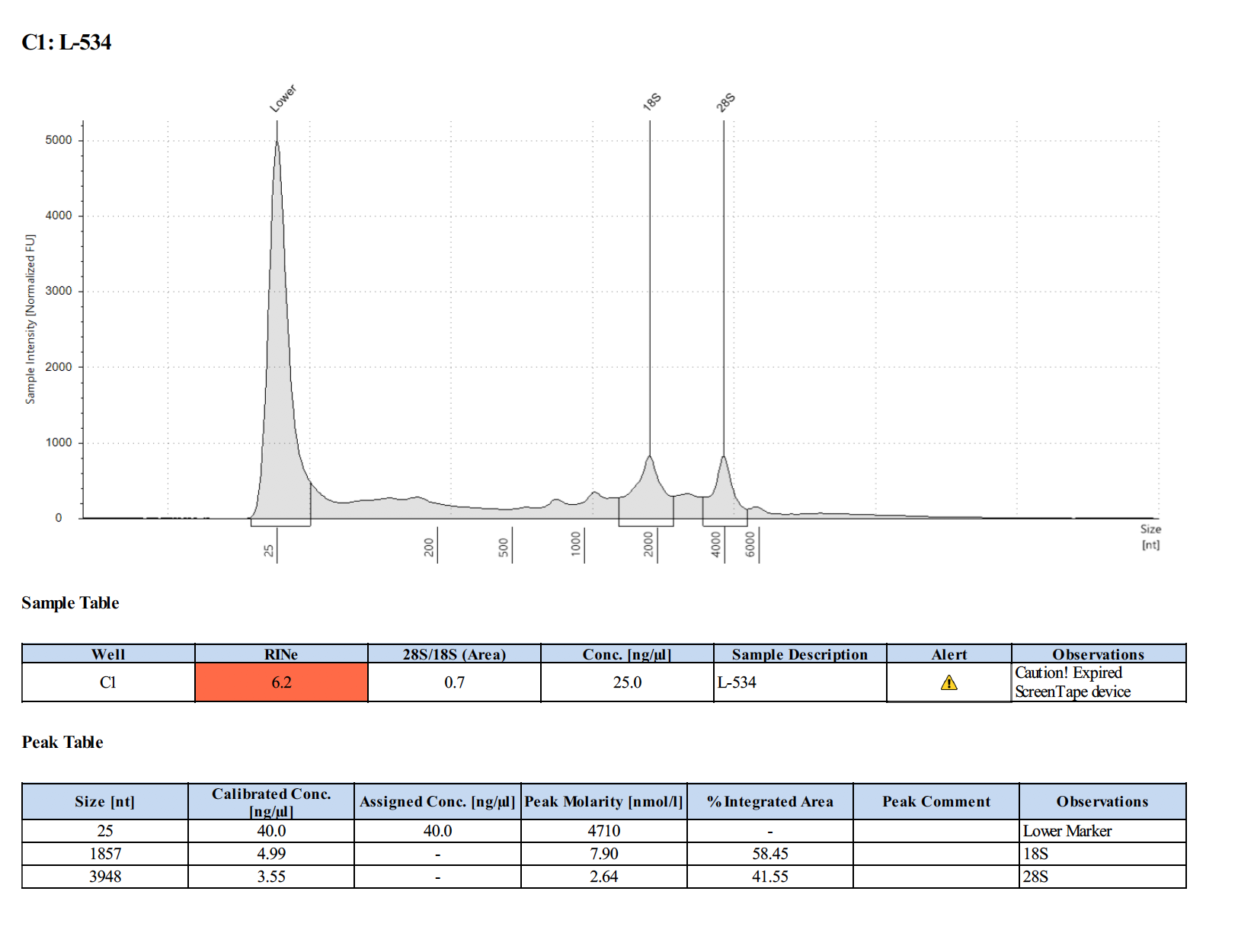
| Larvae | L-534 | 11.4 | 26.8 |
| Larvae | L-939 | 11.3 | 26.6 |
| Larvae | L-1026 | 10.6 | 25.8 |
| Fragment | 18-31 | 4.48 | LOW |
| Fragment | 18-43 | 7.08 | 13.6 |
| Fragment | 18-57 | 29.0 | 36.4 |
Gel Electrophoresis
To test DNA quality: Gel Electrophoresis
- Today I did a small gel of the above protocol and ran it at 80V.

TapeStation
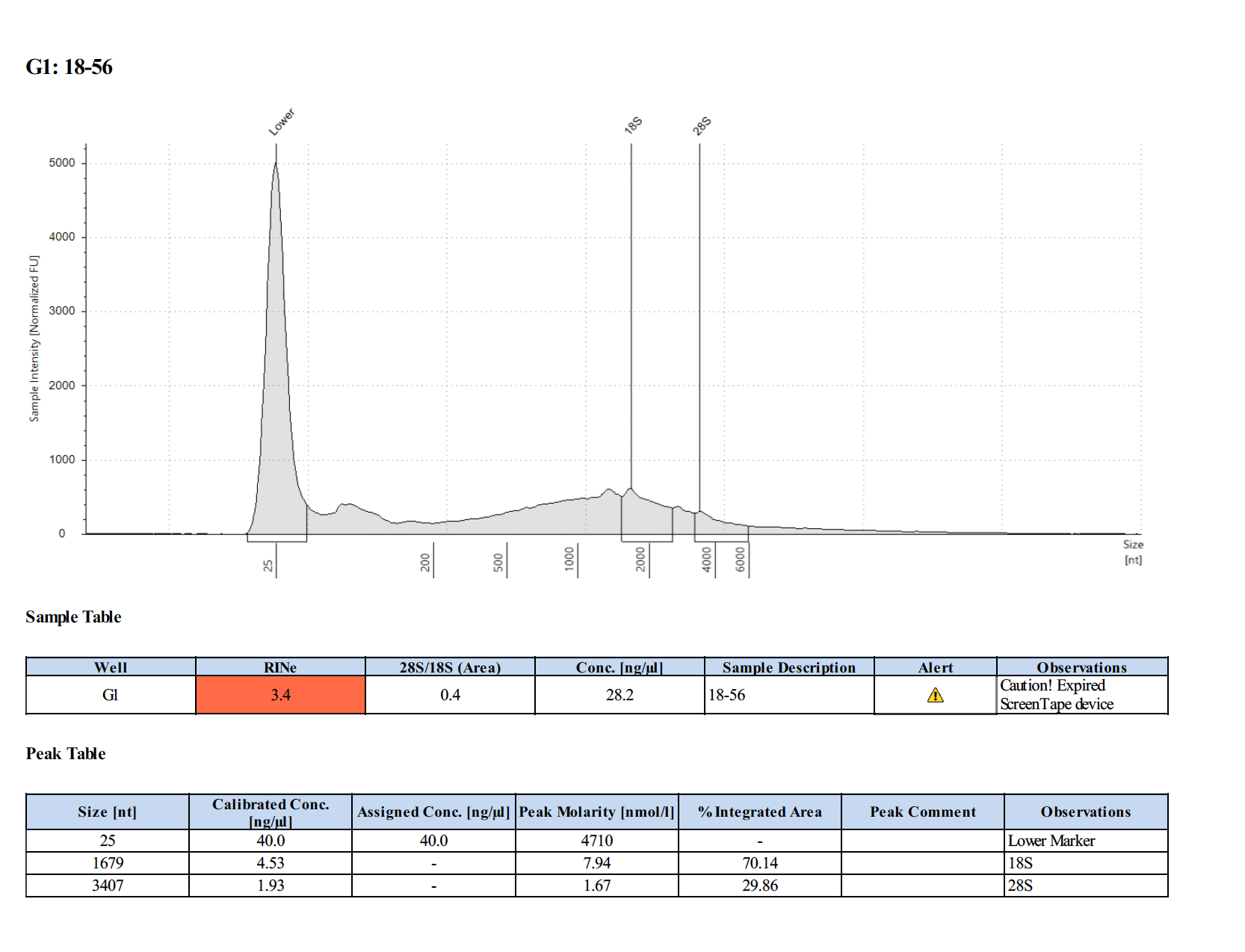
To test RNA quality: TapeStation
Overall Summary:
Sample #17-215:
Sample #1L-534:
Sample #L-939:
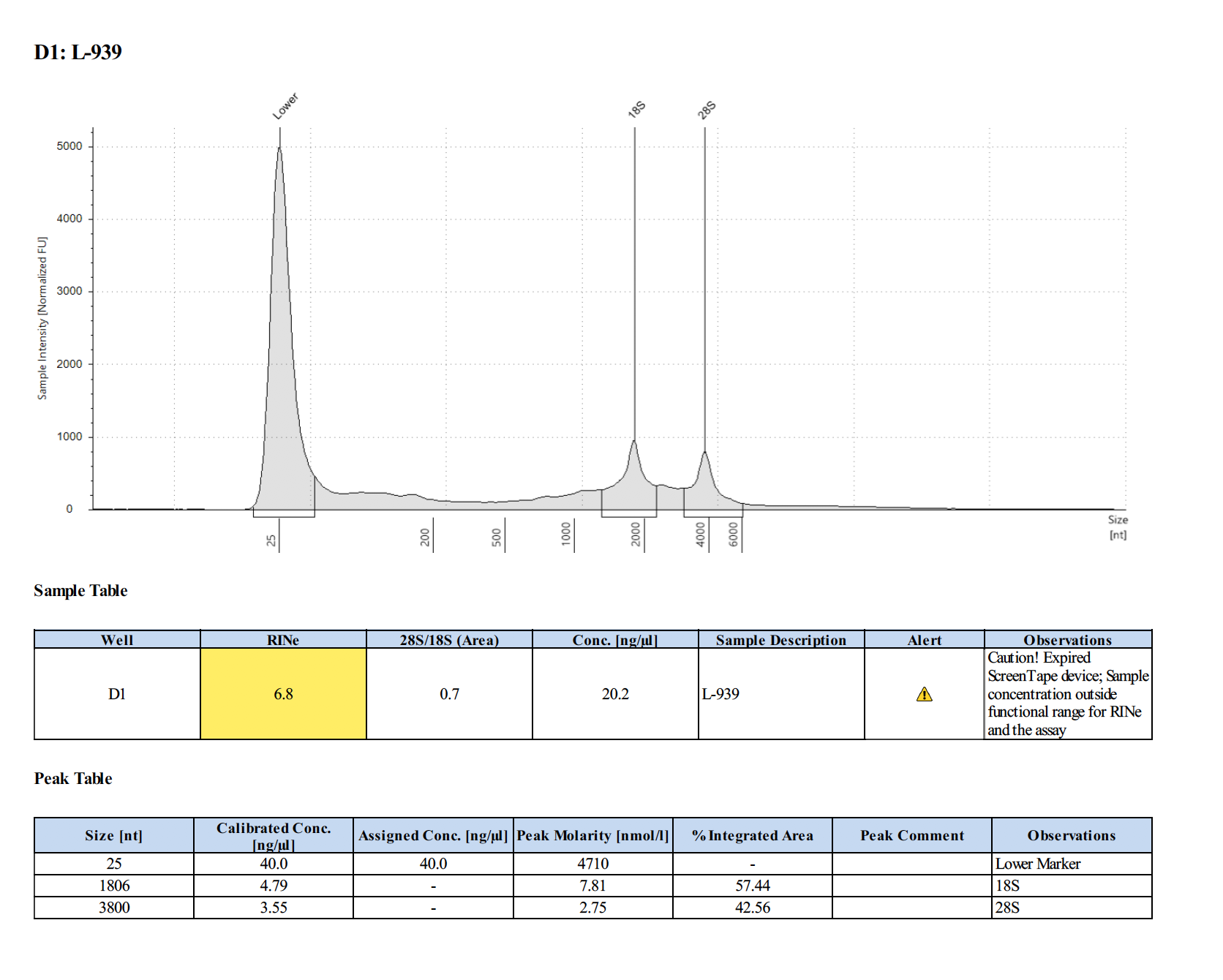
Sample #L-1026:
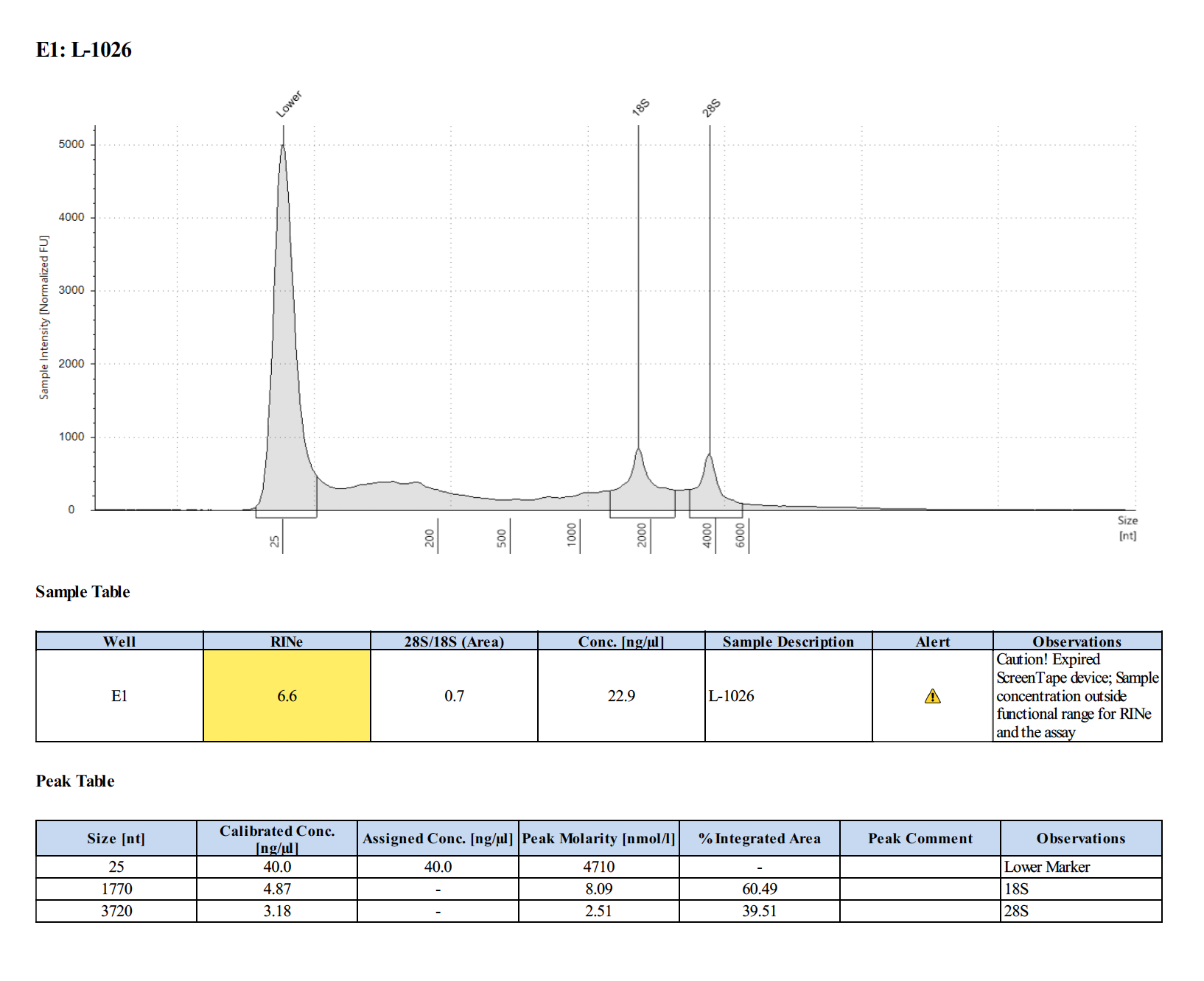
Sample #18-43:
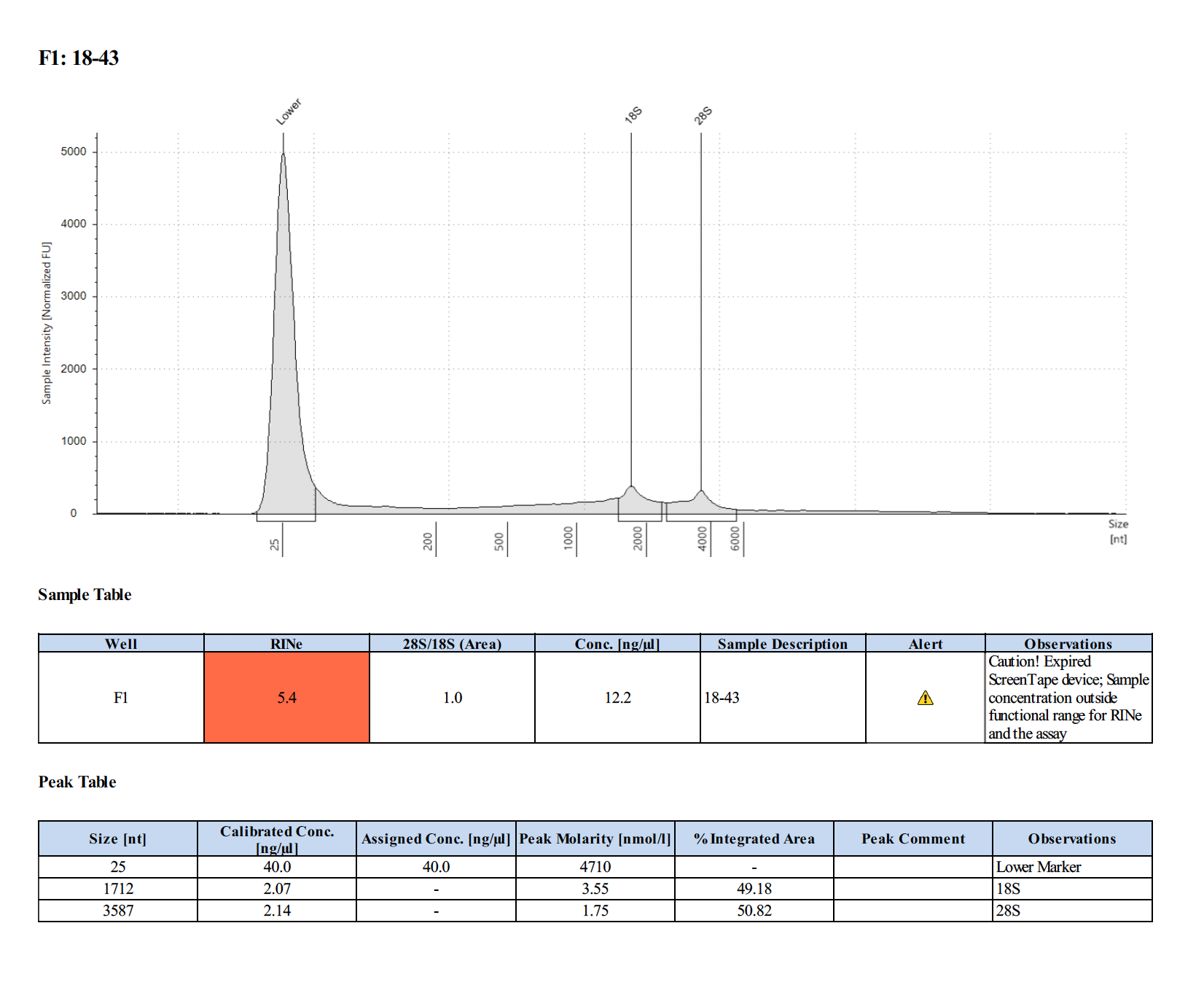
Sample #18-57: