DNA-RNA-Extractions-Thermal-Transplant-Porites-and-Geoduck
Goal
To stream line the protocol for DNA/RNA extractions for Porites astreoides for the Thermal Transplant 2018 experiment and to test larval geoduk extractions for S. Gurr.
Samples
To start, I am using 3 adult P. astreoides samples that were already chipped (~0.5mm diameter) and placed in 500 uL of DNA/RNA Shield, and 2 larval geoducks samples snap frozen in liquid nitrogen.
Sample Numbers
- 55
- Coral: P3-B
- 11
- Coral: R8-B
- 35
- Coral: P12-A
- 618
- Geoduck
- 694
- Geoduck
Protocol description
I used a modified version of this protocol.
Reagent Preparation
- Add 96 mL 100% ethanol (104 mL 95% ethanol) to the 24 mL DNA/RNA Wash Buffer concentrate before use. DNA/RNA Wash Buffer included with D7003T (Mini Prep Plus Kit) is supplied ready-to-use and does not require the addition of ethanol prior to use. Check kit contents and instructions to confirm prep steps.
- Make new 100% Ethanol today
Coral Homogenization
- Thaw samples from -80 °C.
- Add 500 μl of RNA/DNA shield to make the total volume 1 mL
- Samples already had beads in it so I did not add any more beads. (Modify next time)
- Vortex for 2 minutes. Leave the settings on and on max power.
- Remove supernatant from the bead tube and place in a new 1.5 microcentrifuge tube labeled on the side with the extraction sample number and today’s date. Label the cap of the microcentrifuge tube with the sample number. This supernatant will become the sample for “soft homogenization”.
- Add 700 μl of sample, 70 μl of Proteinase K digestion buffer (10:1 ratio of sample:digestion buffer), and 35 μl of Proteinase K (2:1 ratio of digestion buffer:Proteinase K) to a new 1.5 mL microcentrifuge tube.
- Vortex and spin down all tubes.
- Trasfer 800 μl of superatant to a new 5 mL tube.
Geoduck Homogenization
- Removed samples from -80 °C.
- Add 1 mL of RNA/DNA shield
- Add 0.25 mL of 0.5mm glass beads to each tube
- Vortex for 2 minutes. Leave the settings on and on max power.
- Note
- After 2 minutes of vortexing, sample 618 had a broken open shell. Sample 694 had no visible signs of shell or tissue. Sample 618 seemed to provide better extraction yields (see below)
- Remove supernatant from the bead tube and place in a new 1.5 microcentrifuge tube labeled on the side with the extraction sample number and today’s date. Label the cap of the microcentrifuge tube with the sample number. This supernatant will become the sample for “soft homogenization”.
- Add 700 μl of sample, 70 μl of Proteinase K digestion buffer (10:1 ratio of sample:digestion buffer), and 35 μl of Proteinase K (2:1 ratio of digestion buffer:Proteinase K) to a new 1.5 mL microcentrifuge tube.
- Vortex and spin down all tubes.
- Trasfer 800 μl of superatant to a new 5 mL tube.
Zymo Duet RNA DNA Extractions
Modified from the Zymo protocol.
DNA Extraction
- Set up yellow DNA spin columns and collection tubes, label appropriately
- Warm elution liquids to 70 °C (10mM Tris HCl pH. 8.0 and RNase free water)
- Add equal volume (to supernatant; 800 µl) DNA/RNA lysis buffer to each sample tube
- Finger flick to mix tubes
- Add 700 µl (total volume) of sample gently to the yellow DNA spin column
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Important Save the flow through from this step: transfer to a new 5 mL tube labeled for RNA
- Repeat steps 5-7 until all volume is gone. (i.e. x 2)
- Total volume for RNA was 1400 µl
- Add 400 µl DNA/RNA Prep Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 700 µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 400 µl DNA/RNA Wash Buffer genetly to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 2 minutes
- Discard flow through (Zymo kit waste)
- Transfer yellow columns to new 1.5 mL microcentrifuge tubes (“E1”)
- Add 50 µl warmed 10mM Tris HCl to each yellow DNA column by dripping slowly directly on the filter
- Incubate at room temperature for 5 minutes. Centrifuge at 16,000 rcf (g) for 30 seconds keep the flow through
- Add another 50 µl warmed 10mM Tris HCl to each yellow DNA column by dripping slowly directly on the filter
- Incubate at room temperature for 15 minutes.
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Throw away filter and keep flow through.
- Aliquot 10 µl of the elution to 0.5 mL PCR tubes for Qubit and Gel Electrophoresis analysis.
- Store at 4 °C if quantifying the same day or the next, if waiting longer store in -20 °C freezer.
RNA Extraction Can do concurrently with DNA Extraction after DNA Extraction Step 7
- Add equal volume (1400 µl) 100% EtOH to the 5 mL tubes labeled for RNA containing the original yellow column flow through
- Vortex and spin down to mix
- Add 700 µl of that liquid to the green RNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 700 µl to the green RNA spin columns (the rest from the 1.5mL RNA tubes)
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Repeat until all volume is gone (i.e. x 4)
- Get DNase I from freezer
- Discard flow through (Zymo kit waste)
- Add 400 µl DNA/RNA Wash Buffer gently to each green RNA column
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Make DNase I treatment master mix:
- 75µl DNA Digestion buffer x # of samples
- 5µl DNase I x # of samples
Today I had 5 samples, therefore: 375 µl of DNA Digestion buffer and 25 µl of DNase I
- Add 80 µl DNase I treatment master mix directly to the filter of the green RNA columns
- Incubate at room temp for 15 minutes
- Add 400 µl DNA/RNA Prep Buffer gently to each column
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 700 µl DNA/RNA Wash Buffer gently to the green RNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 400 µl DNA/RNA Wash Buffer genetly to the green RNA spin columns
- Centrifuge at 16,000 rcf (g) for 2 minutes
- Discard flow through (Zymo kit waste)
- Transfer green columns to new 1.5 mL microcentrifuge tubes
- Add 50 µl warmed DNase/RNase free water to each green RNA column by dripping slowly directly on the filter
- Incubate at room temp for 5 minutes
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Repeat steps 25-27 for a final elution volume of 100 µl
- Aliquot 5 µl of the final elution to 0.5 mL PCR tubes for Qubit and TapeStation analysis.
- Store all tubes in the -80 °C freezer.
Clean-up
- Place tissue and liquid in the waste container labeled Zymo extraction waste.
- Wipe down RNA free area with RNase away and kimwipes.
- Throw away all tips and restock tip boxes if necessary.
Testing Quantity and Quality
To test RNA and DNA quantity: Qubit
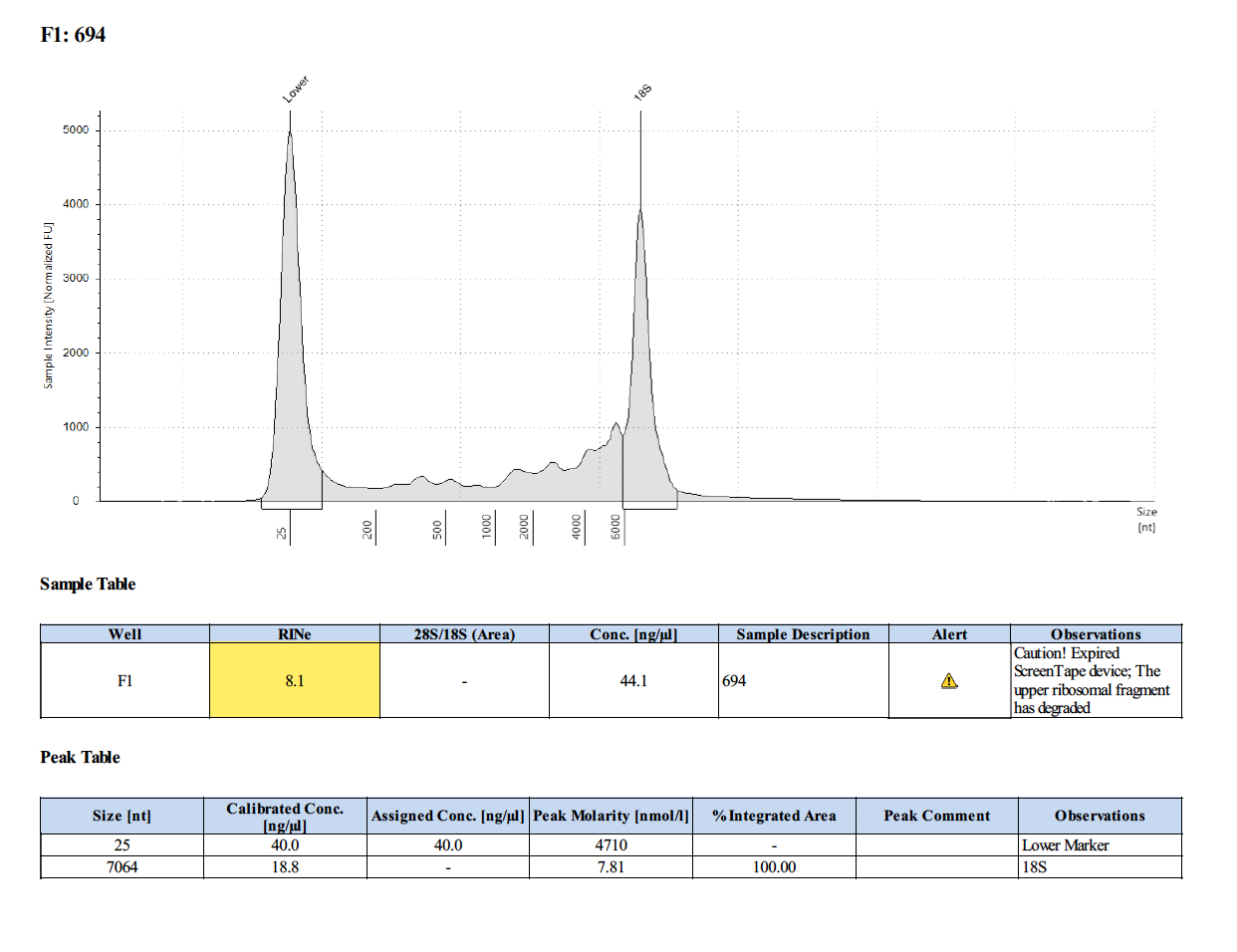
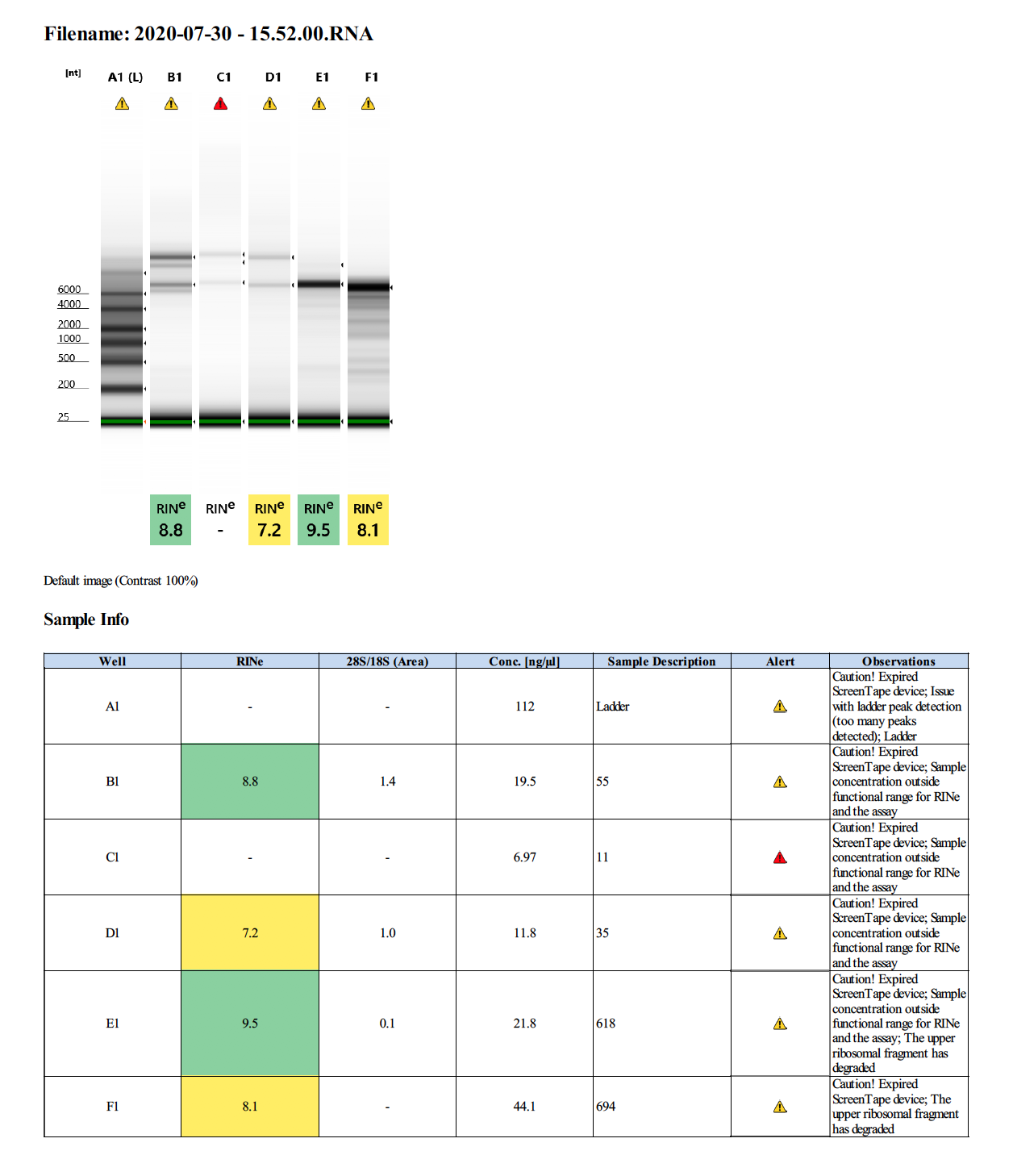
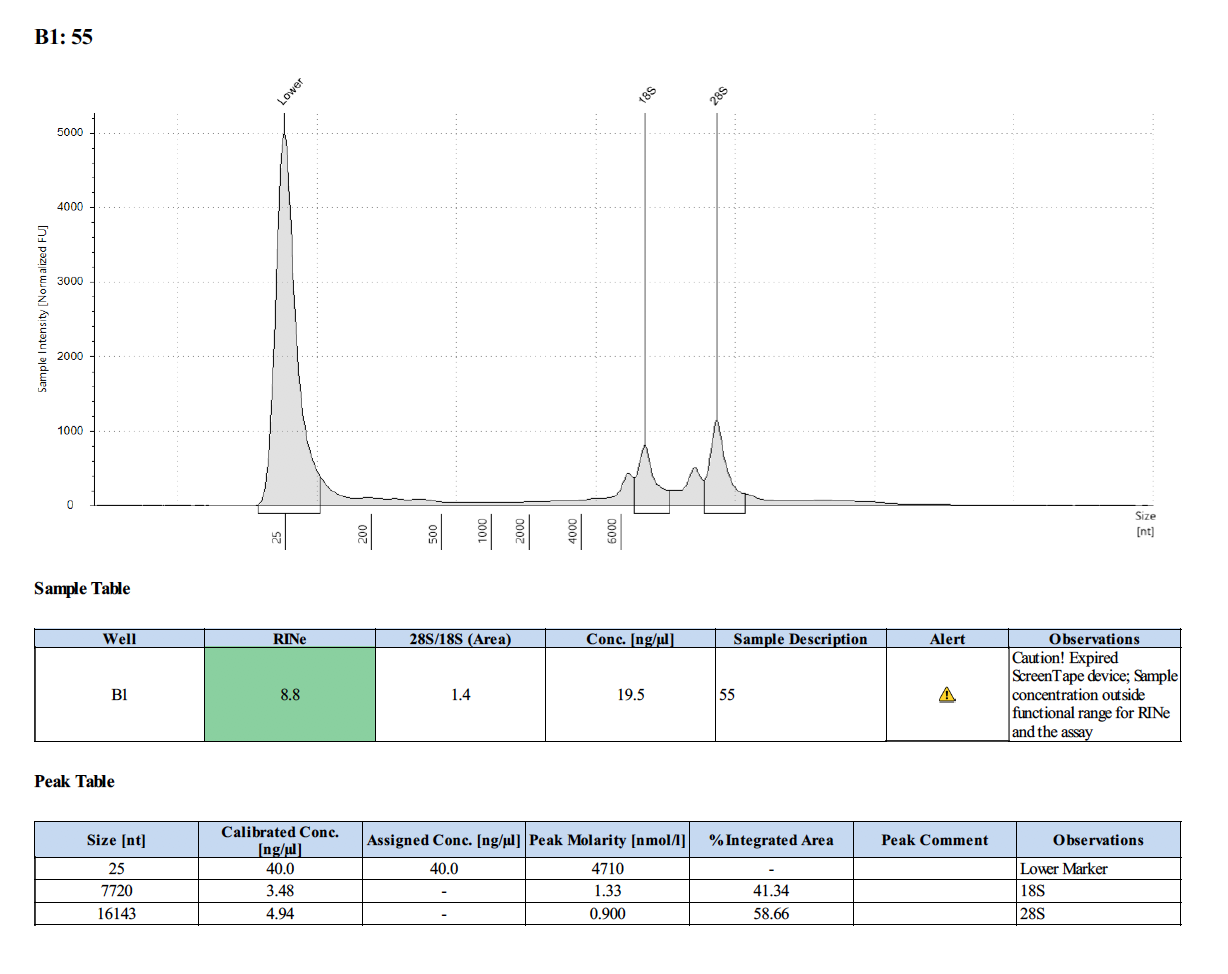
To test RNA quality: TapeStation
To test DNA quality: Gel Electrophoresis
Quantify Results
Qubit
To test RNA and DNA quantity: Qubit
5 samples + 2 standards + 1 error = 8
DNA Broad Range
199 µl Buffer x 8 = 1592 µl Buffer 1 µl Reagent x 8 = 8 µl Reagent
RNA Broad Range
199 µl Buffer x 8 = 1592 µl Buffer 1 µl Reagent x 8 = 8 µl Reagent
| Sample | DNA (ng/uL) | RNA (ng/uL) |
|---|---|---|
| Standard 1 | 155.10 | 425.3 |
| Standard 2 | 23627.15 | 10321.47 |
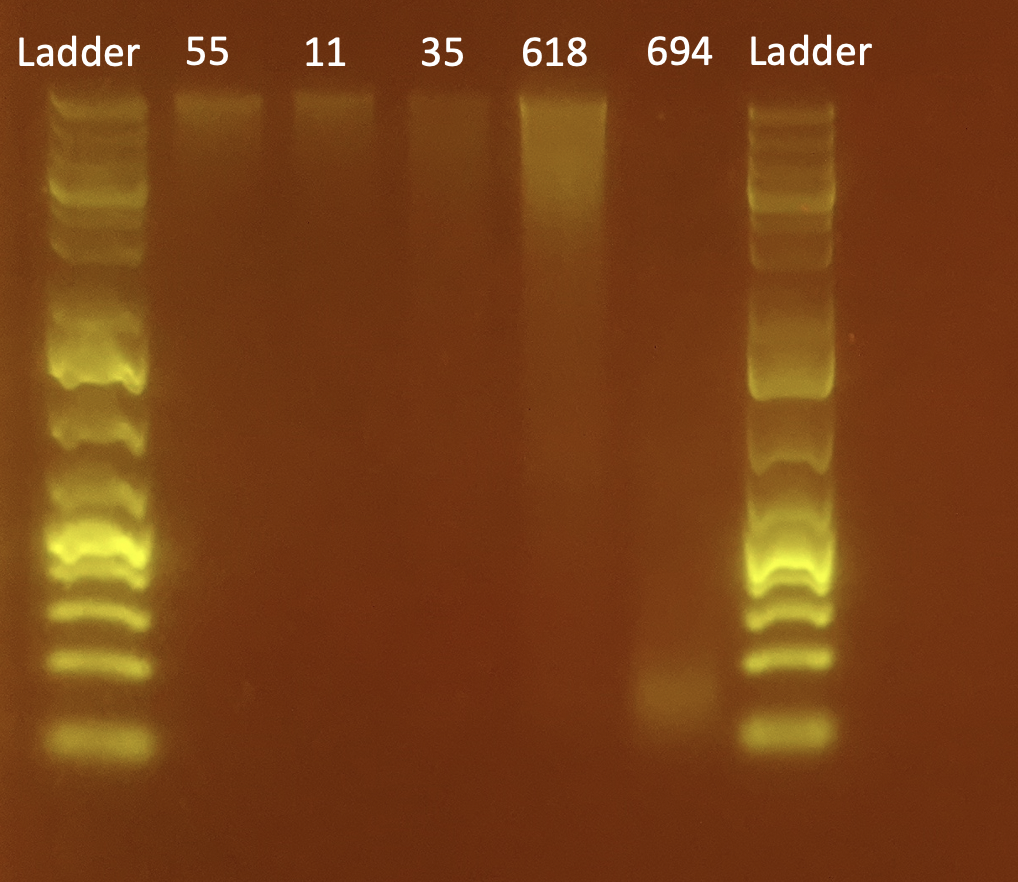
| 55 | 3.98 | 17.0 |
| 11 | LOW | LOW |
| 35 | 2.64 | 13.2 |
| 618 | 20.4 | 22.4 |
| 694 | 5.54 | 62.6 |
Gel Electrophoresis
To test DNA quality: Gel Electrophoresis
- Today I did a small gel of the above protocol
TapeStation
To test RNA quality: TapeStation
Overall Summary:
Sample #55:
Sample #11:
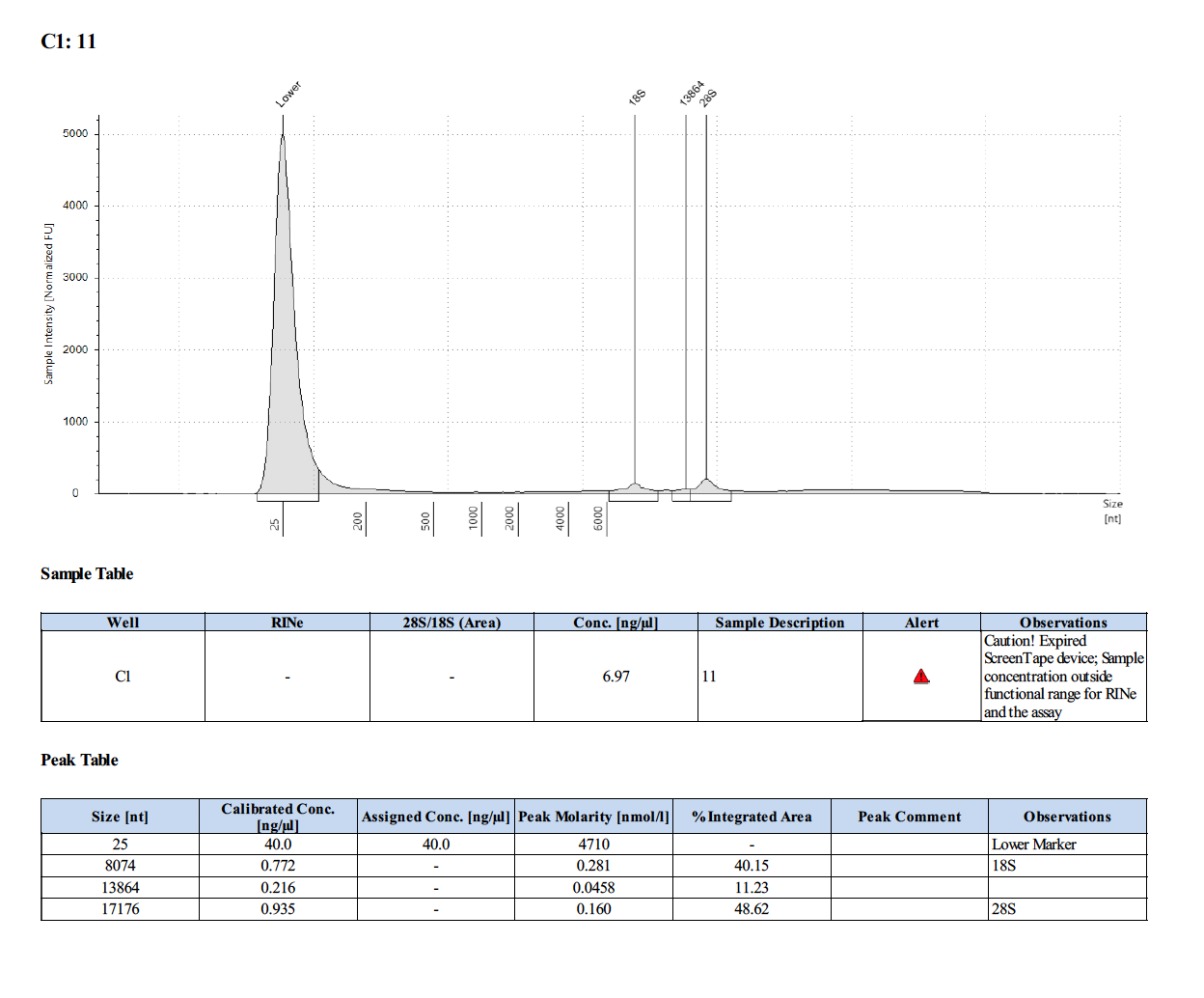
Sample #35:
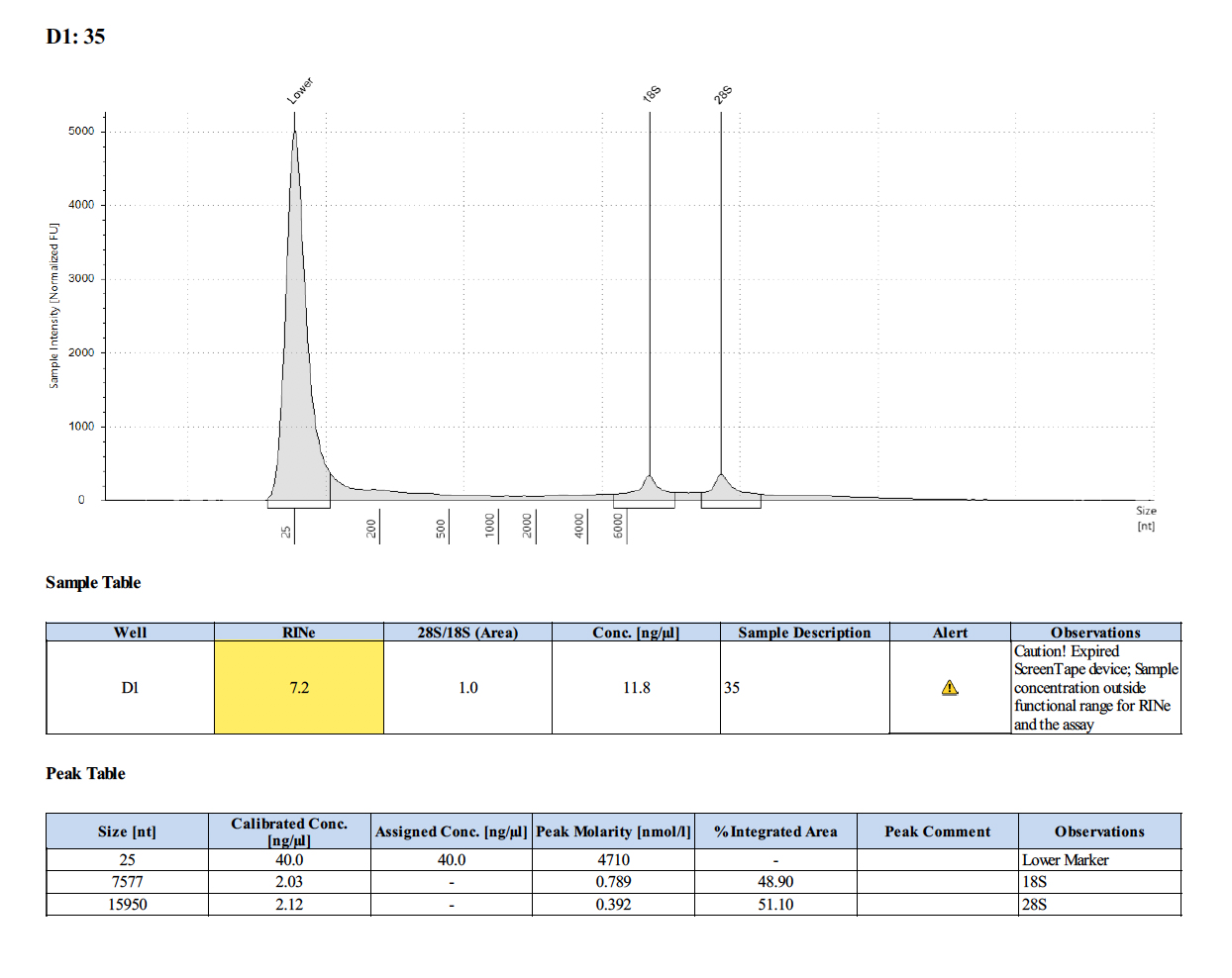
Sample #618:
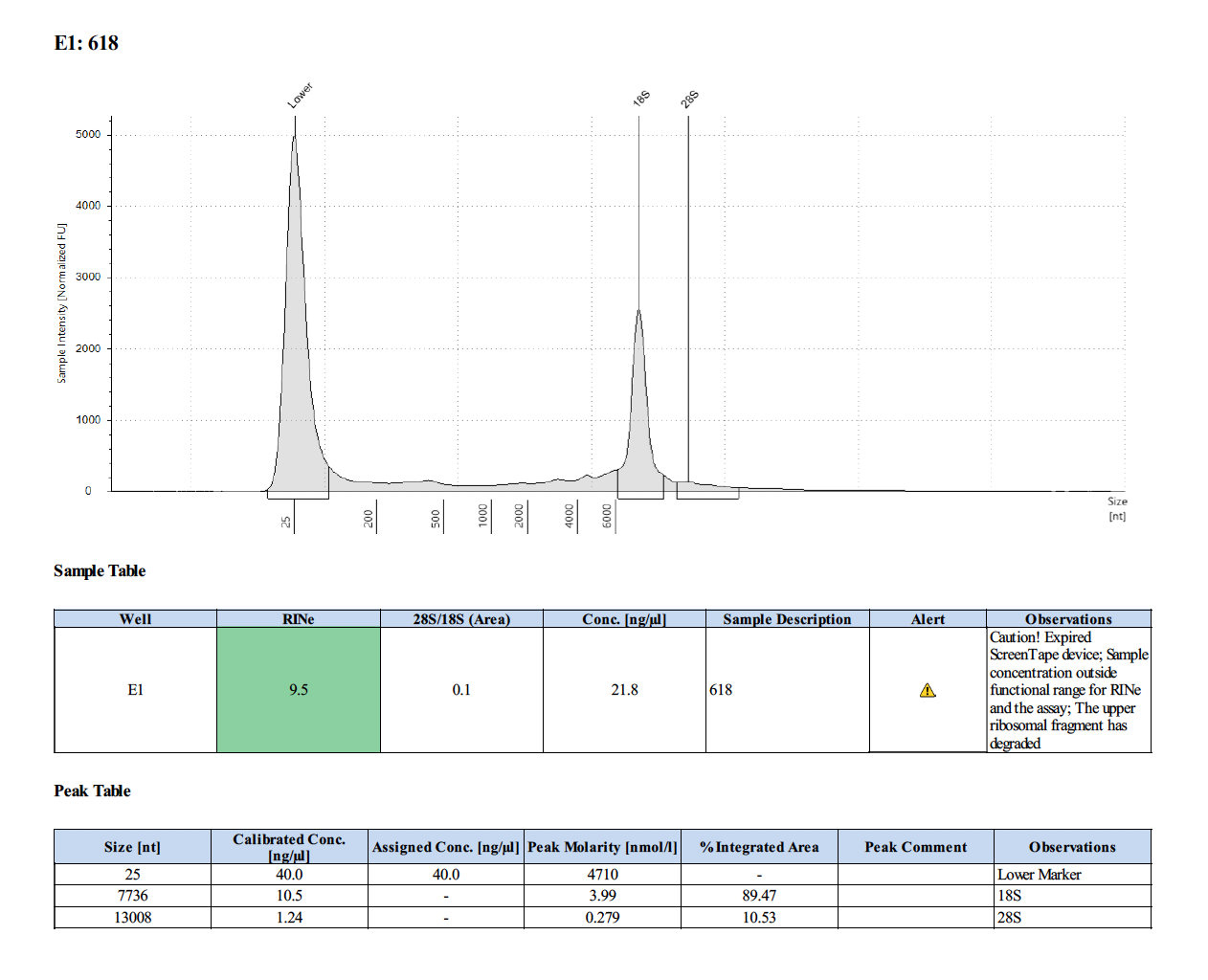
Sample #694: