Direct RNA sequencing on Oxford Nanopore MinION Protocol
Table of Contents
- Overview of Protocol
- Equipment and consumables
- Computer requirements and software
- Library preparation
- Priming and loading the SpotON flow cell
- Data acquisition and basecalling
- Downstream analysis
- Ending the experiment
Overview of Protocol
Direct RNA Sequencing Kit features
This kit is highly recommended for users who:
- are exploring attributes of native RNA such as modified bases
- would like to remove RT or PCR bias
- have transcripts that are difficult to reverse transcribe
Introduction to the Direct RNA Sequencing protocol
This protocol describes how to carry out sequencing of native RNA using the Direct RNA Sequencing Kit (SQK-RNA002).
Compatibility of this protocol
This protocol should only be used in combination with:
- Direct RNA Sequencing Kit (SQK-RNA002)
- R9.4.1 (FLO-MIN106) flow cells
- Flow Cell Wash Kit (EXP-WSH004)
Steps in the sequencing workflow:
1. Prepare for your experiment
- Extract your RNA, and check its length, quantity and purity. The quality checks performed during the protocol are essential in ensuring experimental success.
- Ensure you have your sequencing kit, the correct equipment and third-party reagents
- Download the software for acquiring and analysing your data
- Check your flow cell(s) to ensure it has enough pores for a good sequencing run
2. Library preparation
- Synthesise the complementary strand of the RNA
- Attach sequencing adapters supplied in the kit to the ends of the RNA-cDNA hybrid
- Prime the flow cell, and load your RNA library into the flow cell

3. Sequencing and analysis
- Start a sequencing run using the MinKNOW software, which will collect raw data from the device and convert it into basecalled reads
Equipment and consumables
Materials
- 50 ng of poly(A)-tailed RNA or 500 ng of total RNA in 9 μl
- Direct RNA Sequencing Kit (SQK-RNA002)
- Flow Cell Priming Kit (EXP-FLP002)
Consumables
- 1.5 ml Eppendorf DNA LoBind tubes
- 0.2 ml thin-walled PCR tubes
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- Freshly prepared 70% ethanol in nuclease-free water
- SuperScript™ III Reverse Transcriptase (Thermo Fisher Scientific, 18080044)
- 10 mM dNTP solution (e.g. NEB N0447)
- NEBNext® Quick Ligation Reaction Buffer (NEB, B6058)
- T4 DNA Ligase 2M U/ml (NEB, M0202T/M)
- Agencourt RNAClean XP beads (Beckman Coulter™, A63987)
- Qubit RNA HS Assay Kit (ThermoFisher Q32852)
- Qubit dsDNA HS Assay Kit (ThermoFisher, cat # Q32851)
Equipment
- Hula mixer (gentle rotator mixer)
- Magnetic rack, suitable for 1.5 ml Eppendorf tubes
- Microfuge
- Vortex mixer
- Ice bucket with ice
- Timer
- Thermal cycler
- Qubit fluorometer (or equivalent for QC check)
- P1000 pipette and tips
- P200 pipette and tips
- P100 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- P2 pipette and tips
- Agilent Bioanalyzer (or equivalent; optional)
- Eppendorf 5424 centrifuge (or equivalent; optional)


Computer requirements and software
MinION Mk1C IT requirements
The MinION Mk1C contains fully-integrated compute and screen, removing the need for any accessories to generate and analyse nanopore data.
Software for nanopore sequencing
The MinKNOW software controls the nanopore sequencing device, collects sequencing data and basecalls in real time. You will be using MinKNOW for every sequencing experiment to sequence, basecall and demultiplex if your samples were barcoded.
Library preparation
Time: ~115 Minutes
Materials
- 50 ng of poly(A)-tailed RNA or 500 ng of total RNA in 9 μl
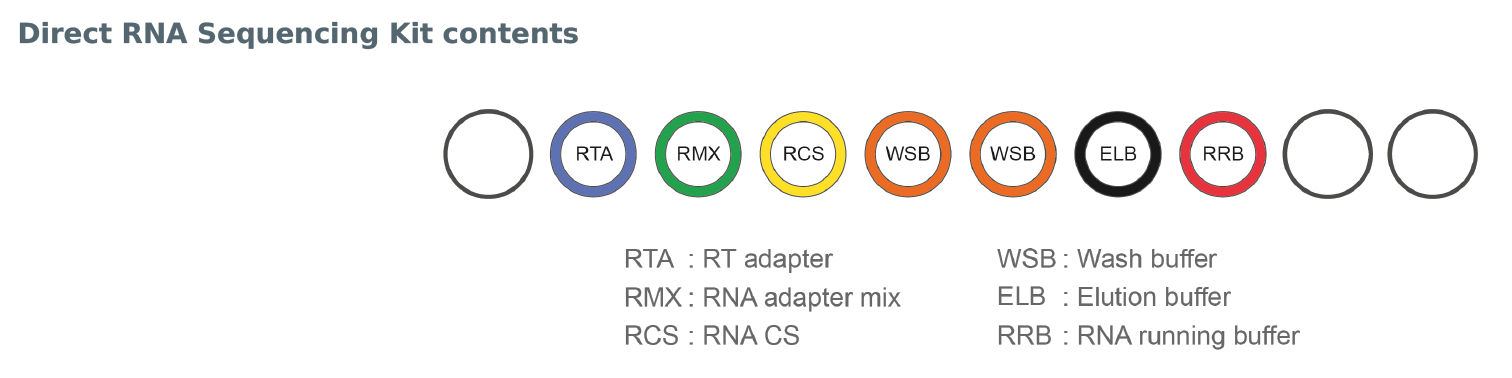
- RT Adapter (RTA)
- RNA CS (RCS)
- RNA Adapter (RMX)
- Wash Buffer (WSB)
- Elution Buffer (ELB)
Consumables
- NEBNext® Quick Ligation Reaction Buffer (NEB, B6058)
- T4 DNA Ligase 2M U/ml (NEB, M0202T/M)
- 0.2 ml thin-walled PCR tubes
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- Agencourt RNAClean XP beads (Beckman Coulter™, A63987)
- Freshly prepared 70% ethanol in nuclease-free water
- 1.5 ml Eppendorf DNA LoBind tubes
- SuperScript™ III Reverse Transcriptase (Thermo Fisher Scientific, 18080044)
- 10 mM dNTP solution (e.g. NEB N0447)
- Qubit dsDNA HS Assay Kit (ThermoFisher, cat # Q32851)
Equipment
- Magnetic rack, suitable for 1.5 ml Eppendorf tubes
- Hula mixer (gentle rotator mixer)
- Thermal cycler
- Qubit fluorometer (or equivalent for QC check)
Protocol
- Prepare the RNA in nuclease-free water.
- Transfer 50 ng of poly(A)-tailed RNA or 500 ng of total RNA into a 1.5 ml Eppendorf DNA LoBind tube
- Adjust the volume to 9 μl with nuclease-free water
- Mix thoroughly by flicking the tube to avoid unwanted shearing
- Spin down briefly in a microfuge
- Spin down all reagents and pipette mix. In a 0.2 ml thin-walled PCR tube, mix the reagents in the following order:
| Reagent | Volume (μl) |
|---|---|
| NEBNext Quick Ligation Reaction Buffer | 3.0 |
| RNA | 9.0 |
| RNA CS (RCS), 110 nM | 0.5 |
| RT Adapter (RTA) | 1.0 |
| T4 DNA Ligase | 1.5 |
| Total | 15 |
- Mix by pipetting and spin down.
- Incubate the reaction for 10 minutes at room temperature.
- Mix the following reagents together to make the reverse transcription master mix:
| Reagent Volume | Volume (μl) |
|---|---|
| Nuclease-free water 9.0 | 9.0 |
| 10 mM dNTPs 2.0 μl | 2.0 |
| 5x first-strand buffer | 8.0 |
| 0.1 M DTT | 4.0 |
| Total | 23.0 |
- Add the master mix to the 0.2 ml PCR tube containing the RT adapter-ligated RNA from the “RT Adapter ligation” step. Mix by pipetting.
- Add 2 μl of SuperScript III Reverse Transcriptase to the reaction and mix by pipetting.
- Place the tube in a thermal cycler and incubate at 50°C for 50 minutes, then 70°C for 10 minutes, and bring the sample to 4°C before proceeding to the next step.
- Transfer the sample to a clean 1.5 ml Eppendorf DNA LoBind tube.
- Resuspend the stock of Agencourt RNAClean XP beads by vortexing.
- Add 72 μl of resuspended Agencourt RNAClean XP beads to the reverse transcription reaction and mix by pipetting.
- Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
- Prepare 200 μl of fresh 70% ethanol in nuclease-free water.
- Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
- Keep the tube on magnet, and wash the beads with 150 μl of freshly prepared 70% ethanol without disturbing the pellet as described below.
- Keeping the magnetic rack on the benchtop, rotate the bead-containing tube by 180°. Wait for the beads to migrate towards the magnet and form a pellet.
- Rotate the tube 180° again (back to the starting position), and wait for the beads to pellet.
- Remove the 70% ethanol using a pipette and discard.
- Spin down and place the tube back on the magnet until the eluate is clear and colourless. Keep the tubes on the magnet and pipette off any residual ethanol.
- Remove the tube from the magnetic rack and resuspend the pellet in 20 μl nuclease-free water. Incubate for 5 minutes at room temperature.
- Pellet the beads on a magnet until the eluate is clear and colourless.
- Remove and retain 20 μl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
- In the same 1.5 ml Eppendorf DNA LoBind tube, mix the reagents in the following order:
| Reagent | Volume (μl) |
|---|---|
| NEBNext Quick Ligation Reaction Buffer | 8.0 |
| RNA Adapter (RMX) | 6.0 |
| Nuclease-free water | 3.0 |
| T4 DNA Ligase | 3.0 |
| Total (including all reagents) | 40.0 |
- Mix by pipetting.
- Incubate the reaction for 10 minutes at room temperature.
- Resuspend the stock of Agencourt RNAClean XP beads by vortexing.
- Add 16 μl of resuspended Agencourt RNAClean XP beads to the reaction and mix by pipetting.
- Incubate on a Hula mixer (rotator mixer) for 5 minutes at room temperature.
- Spin down the sample and pellet on a magnet. Keep the tube on the magnet, and pipette off the supernatant when clear and colourless.
- Add 150 μl of the Wash Buffer (WSB) to the beads. Close the tube lid and resuspend the beads by flicking the tube.Return the tube to the magnetic rack, allow the beads to pellet for 5 minutes and pipette off the supernatant it is 29. when clear and colourless.
- Repeat the previous step.
- Important: Agitating the beads results in a more efficient removal of free adapter, compared to adding the wash buffer and immediately aspirating.
- Remove the tube from the magnetic rack and resuspend pellet in 21 μl Elution Buffer by the gently flicking the tube. Incubate for 10 minutes at room temperature.
- Pellet the beads on a magnet until the eluate is clear and colourless.
- Remove and retain 21 μl of eluate into a clean 1.5 ml Eppendorf DNA LoBind tube.
- Quantify 1 μl of reverse-transcribed and adapted RNA using the Qubit fluorometer DNA HS assay - recovery aim ~20 ng.
Priming and loading the SpotON flow cell
Time: ~15 minutes
Materials
- Prepared RNA library
- RNA Running Buffer (RRB)
- Flow Cell Priming Kit (EXP-FLP002)
Consumables
- MinION Mk1B
- SpotON Flow Cell
- Nuclease-free water (e.g. ThermoFisher, AM9937)
- 1.5 ml Eppendorf DNA LoBind tubes
Protocol
- Thaw the RNA Running Buffer (RRB), Flush Tether (FLT) and one tube of Flush Buffer (FB) at room temperature.
- Mix the RNA Running Buffer (RRB), Flush Buffer (FB) and Flush Tether (FLT) tubes thoroughly by vortexing and spin down at room temperature.
- To prepare the flow cell priming mix, add 30 μl of thawed and mixed Flush Tether (FLT) directly to the tube of thawed and mixed Flush Buffer (FB), and mix by vortexing at room temperature.
- Open the MinION device lid and slide the flow cell under the clip.
- Press down firmly on the flow cell to ensure correct thermal and electrical contact.
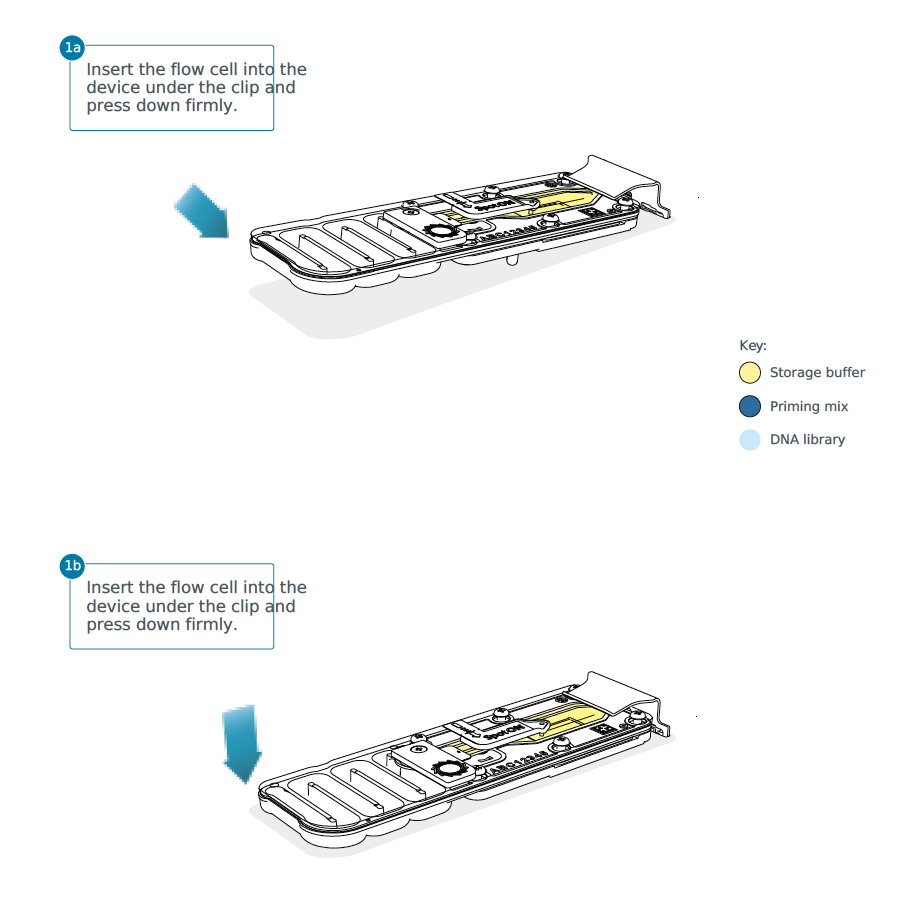
- Slide the priming port cover clockwise to open the priming port.
- After opening the priming port, check for a small air bubble under the cover. Draw back a small volume to remove any bubbles:
- Set a P1000 pipette to 200 μl
- Insert the tip into the priming port
- Turn the wheel until the dial shows 220-230 ul, to draw back 20-30 ul, or until you can see a small volume of buffer entering the pipette tip
- Note: Visually check that there is continuous buffer from the priming port across the sensor array.

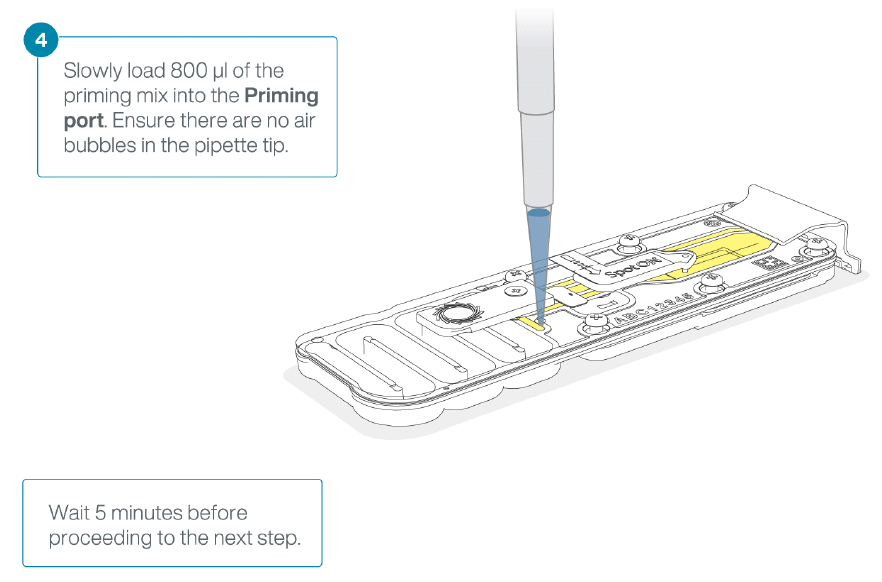
- Load 800 μl of the priming mix into the flow cell via the priming port, avoiding the introduction of air bubbles. Wait for five minutes. During this time, prepare the library for loading by following the steps below.
- Take 20 μl of the prepared RNA library and mix it with 17.5 μl of nuclease-free water.
- In a new tube, prepare the library for loading as follows:
| Reagent | Volume per flow cell (μl) |
|---|---|
| RNA Running Buffer (RRB) | 37.5 |
| RNA library in nuclease-free water | 37.5 |
| Total | 75 |
- Complete the flow cell priming:
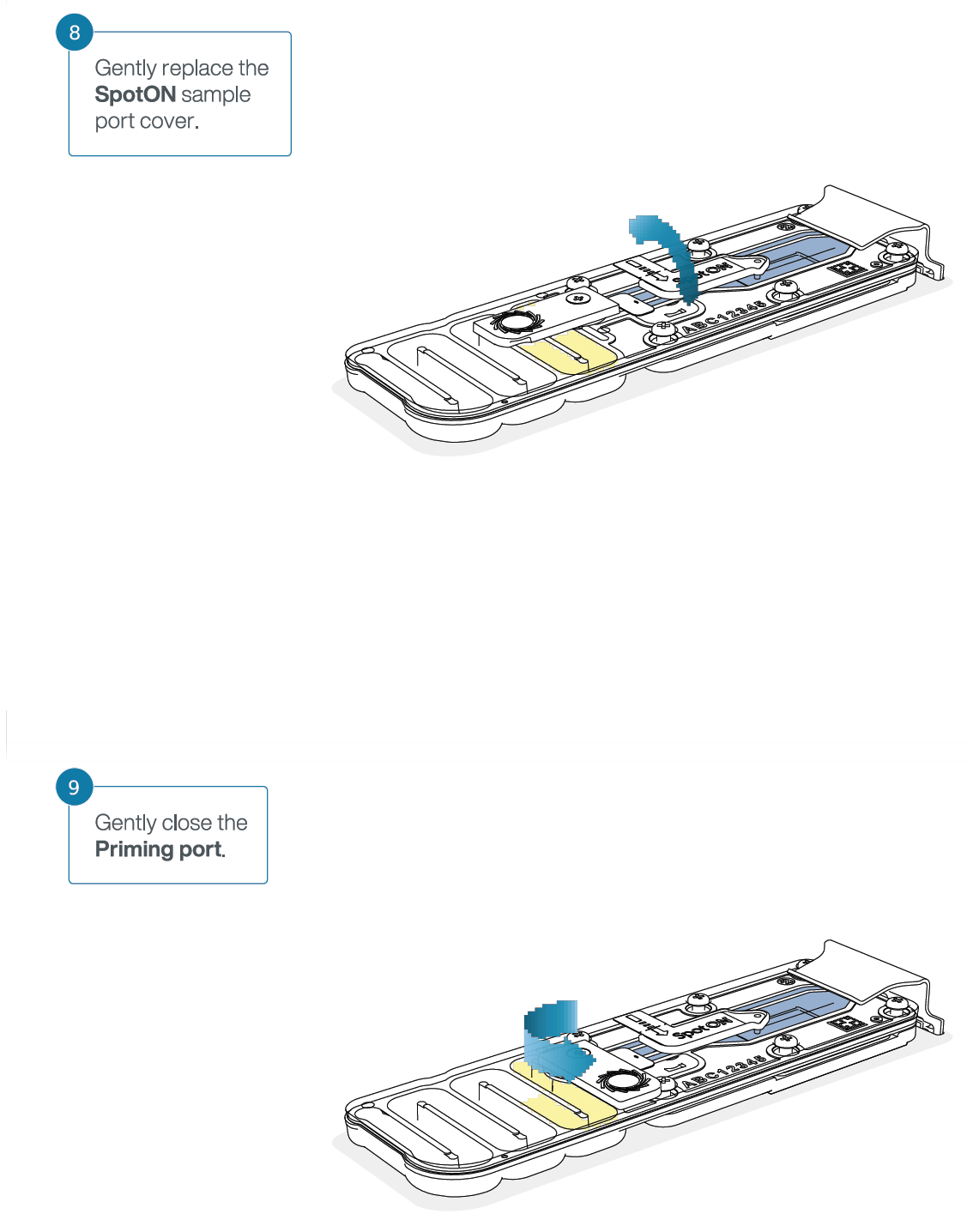
- Gently lift the SpotON sample port cover to make the SpotON sample port accessible.
- Load 200 μl of the priming mix into the flow cell priming port (not the SpotON sample port), avoiding the introduction of air bubbles.

- Mix the prepared library gently by pipetting up and down just prior to loading.
- Add 75 μl of sample to the Flow Cell via the SpotON sample port in a dropwise fashion. Ensure each drop flows into the port before adding the next.

- Gently replace the SpotON sample port cover, making sure the bung enters the SpotON port, close the priming port and replace the MinION device lid.