Gfas Dark Genes Single cell preliminary analysis
Goal:
- To assess the quality of our single cell data on a control (ambient) sample
Data:
- Single cell suspension prepartion protocol (add)
- This is coral W-045
- From the 10X Chromium sequencing (10X 3’ v3.1), we performed shallow sequencing (500 reads per cell)
- This produced 10 files
Reference Genomes:
- Galaxea fasicularis v1
- From Reef Genomics
- Download site
- Durisdinum trenchii (SCF082)
Transfer data from Box to Pegasus
-
I manually downloaded the files from Box to my hard drive.
-
md5 on hard drive files
All Cells:
$ cd 20230905_SingleCell_DG/
$ md5 *.gz
MD5 (AndradeRodriguez-15275-001_GEX3_S9_L001_R1_001.fastq.gz) = a41d311fed6542869cbfca5d933a3d5d
MD5 (AndradeRodriguez-15275-001_GEX3_S9_L001_R2_001.fastq.gz) = 8a70aaa04c6bffbf2bcb4576f4aeb9b3
MD5 (AndradeRodriguez-15275-001_GEX3_S9_L002_R1_001.fastq.gz) = 06f5e9b6b9b6713fe65376dc53539488
MD5 (AndradeRodriguez-15275-001_GEX3_S9_L002_R2_001.fastq.gz) = 7dcffd1668d4b335061bdaded9d9e898
MD5 (AndradeRodriguez-15275-001_GEX3_S9_L003_R1_001.fastq.gz) = 8e801a9bb82cedac7e6b3470e27e4095
MD5 (AndradeRodriguez-15275-001_GEX3_S9_L003_R2_001.fastq.gz) = 4eff72dca6601f22134982a6c0a3577f
MD5 (AndradeRodriguez-15275-001_GEX3_S9_L004_R1_001.fastq.gz) = f9b85b014e123dcadf854ea037cf7f75
MD5 (AndradeRodriguez-15275-001_GEX3_S9_L004_R2_001.fastq.gz) = b953a5d66ba7b47e19d3485cdc3a774c
MD5 (AndradeRodriguez-15275-001_GEX3_S9_L005_R1_001.fastq.gz) = 3df3e6b965af17f451df16dab1604110
MD5 (AndradeRodriguez-15275-001_GEX3_S9_L005_R2_001.fastq.gz) = 4a33ee1e8dda001a9b1d7463afbe726b
- Transfer data from hard drive to Pegasus
Raw fastq files:
$ scp -r /Volumes/Passport/Kevin/Andrade_Rodriguez-21-07-15275-396479269/20230905_SingleCell_DG kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/
Galaxea fasicularis genome files:
$ scp -r /Volumes/Passport/Kevin/Gfas_v1 kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/
Durisdinum trenchii genome files:
$ scp -r /Volumes/Passport/Kevin/Dtrenchii_SCF082 kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/
- md5 file for Pegasus files
$ cd 20230905_SingleCell_DG/
$ md5sum *.gz
a41d311fed6542869cbfca5d933a3d5d AndradeRodriguez-15275-001_GEX3_S9_L001_R1_001.fastq.gz
8a70aaa04c6bffbf2bcb4576f4aeb9b3 AndradeRodriguez-15275-001_GEX3_S9_L001_R2_001.fastq.gz
06f5e9b6b9b6713fe65376dc53539488 AndradeRodriguez-15275-001_GEX3_S9_L002_R1_001.fastq.gz
7dcffd1668d4b335061bdaded9d9e898 AndradeRodriguez-15275-001_GEX3_S9_L002_R2_001.fastq.gz
8e801a9bb82cedac7e6b3470e27e4095 AndradeRodriguez-15275-001_GEX3_S9_L003_R1_001.fastq.gz
4eff72dca6601f22134982a6c0a3577f AndradeRodriguez-15275-001_GEX3_S9_L003_R2_001.fastq.gz
f9b85b014e123dcadf854ea037cf7f75 AndradeRodriguez-15275-001_GEX3_S9_L004_R1_001.fastq.gz
b953a5d66ba7b47e19d3485cdc3a774c AndradeRodriguez-15275-001_GEX3_S9_L004_R2_001.fastq.gz
3df3e6b965af17f451df16dab1604110 AndradeRodriguez-15275-001_GEX3_S9_L005_R1_001.fastq.gz
4a33ee1e8dda001a9b1d7463afbe726b AndradeRodriguez-15275-001_GEX3_S9_L005_R2_001.fastq.gz
Build a custom reference
- Decompress files
$ gunzip gfas_1.0.genes.gff3.gz
$ gunzip gfas_final_1.0.fasta.gz
I have to convert this GFF3 file to GTF:
$ nano convert.job
#!/bin/bash
#BSUB -J scSeq_convertGTF
#BSUB -q general
#BSUB -P dark_genes
#BSUB -n 6
#BSUB -W 12:00
#BSUB -u kxw755@earth.miami.edu
#BSUB -o gfas_convertGTF.out
#BSUB -e gfas_convertGTF.err
###################################################################
module load cufflinks/2.2.1
gffread gfas_1.0.genes.gff3 -T -o gfas_1.0.gtf
$ bsub < convert.job
- Filter GTF
$ nano mkgtf.job
#!/bin/bash
#BSUB -J scSeq_mkgtf_dg1
#BSUB -q general
#BSUB -P dark_genes
#BSUB -n 6
#BSUB -W 12:00
#BSUB -u kxw755@earth.miami.edu
#BSUB -o gfas_mkgtf_v1.out
#BSUB -e gfas_mkgtf_v1.err
###################################################################
cellranger mkgtf \
gfas_1.0.gtf \
gfas_1.0.filtered.gtf \
--attribute=gene_biotype:protein_coding
$ bsub < mkgtf.job
- Make Reference
$ nano mkref.job
#!/bin/bash
#BSUB -J scSeq_mkref_dg1
#BSUB -q general
#BSUB -P dark_genes
#BSUB -n 6
#BSUB -W 12:00
#BSUB -u kxw755@earth.miami.edu
#BSUB -o gfas_mkref_v1.out
#BSUB -e gfas_mkref_v1.err
###################################################################
cellranger mkref \
--genome=gfas \
--fasta=gfas_final_1.0.fasta \
--genes=gfas_1.0.filtered.gtf
$ bsub < mkref.job
Count
$ cd /nethome/kxw755/20230905_SingleCell_DG/raw_data
$ nano count_W045_1.job
#BSUB -J count_W045
#BSUB -q general
#BSUB -P dark_genes
#BSUB -n 6
#BSUB -W 120:00
#BSUB -u kxw755@earth.miami.edu
#BSUB -o count.out
#BSUB -e count.err
#BSUB -B
#BSUB -N
###################################################################
cellranger count \
--id=W-045_1 \
--transcriptome=/nethome/kxw755/Gfas_v1/gfas \
--fastqs=/nethome/kxw755/20230905_SingleCell_DG/raw_data \
--sample=AndradeRodriguez-15275-001_GEX3
$ bsub < count_W045_1.job
Exporting files
scp -r kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/20230905_SingleCell_DG/raw_data/W-045_1/outs/filtered_feature_bc_matrix.h5 /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/W-045_1_raw_feature_bc_matrix.h5
scp -r kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/20230905_SingleCell_DG/raw_data/W-045_1/outs/web_summary.html /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/W-045_1_web_summary.html
Summary


20231004 Analysis
Since this run went well, we asked Oncogenomics to so a deeper sequencing (50,000 reads per cell) on the same sample (5000 cells)
Transfer data from Box to Pegasus
-
I manually downloaded the files from Box to my hard drive.
-
md5 on hard drive files
All Cells:
Copying files form my computer to Pegasus
scp -r /Users/kevinwong/Desktop/BCL_Convert_10_02_2023_9_01_08-695873424.zip kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/
unzip files
unzip BCL_Convert_10_02_2023_9_01_08-695873424.zip
Make a new directory
mkdir 20231004_SingleCell_DG
Find all *.gz files in lower directories and copy them into one directory
find /BCL_Convert_10_02_2023_9_01_08-695873424 -type f -name "*.gz" -exec mv {} ./20231004_SingleCell_DG \:
This kind of worked, but I ended up using mv to copy them into one foler individually.
There are 16 files in total:
AndradeRodriguez-15275-001_GEX3_S14_L001_R1_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L001_R2_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L003_R1_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L005_R1_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L007_R1_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L003_R2_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L005_R2_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L007_R2_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L002_R1_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L004_R1_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L006_R1_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L008_R1_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L002_R2_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L004_R2_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L006_R2_001.fastq.gz
AndradeRodriguez-15275-001_GEX3_S14_L008_R2_001.fastq.gz
Count
$ cd /nethome/kxw755/20231004_SingleCell_DG
$ nano count_W045_deep.job
#BSUB -J count_W045_deep
#BSUB -q general
#BSUB -P dark_genes
#BSUB -n 6
#BSUB -W 120:00
#BSUB -u kxw755@earth.miami.edu
#BSUB -o count.out
#BSUB -e count.err
#BSUB -B
#BSUB -N
###################################################################
cellranger count \
--id=W-045_1_deep \
--transcriptome=/nethome/kxw755/Gfas_v1/gfas \
--fastqs=/nethome/kxw755/20231004_SingleCell_DG \
--sample=AndradeRodriguez-15275-001_GEX3
$ bsub < count_W045_deep.job
Exporting files
scp -r kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/20231004_SingleCell_DG/W-045_1_deep/outs/filtered_feature_bc_matrix.h5 /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/W-045_1_deep_raw_feature_bc_matrix.h5
scp -r kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/20231004_SingleCell_DG/W-045_1_deep/outs/web_summary.html /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/W-045_1_deep_web_summary.html
scp -r kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/20231004_SingleCell_DG/W-045_1_deep/outs/metrics_summary.csv /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/W-045_1_deep_metrics_summary.csv
Summary
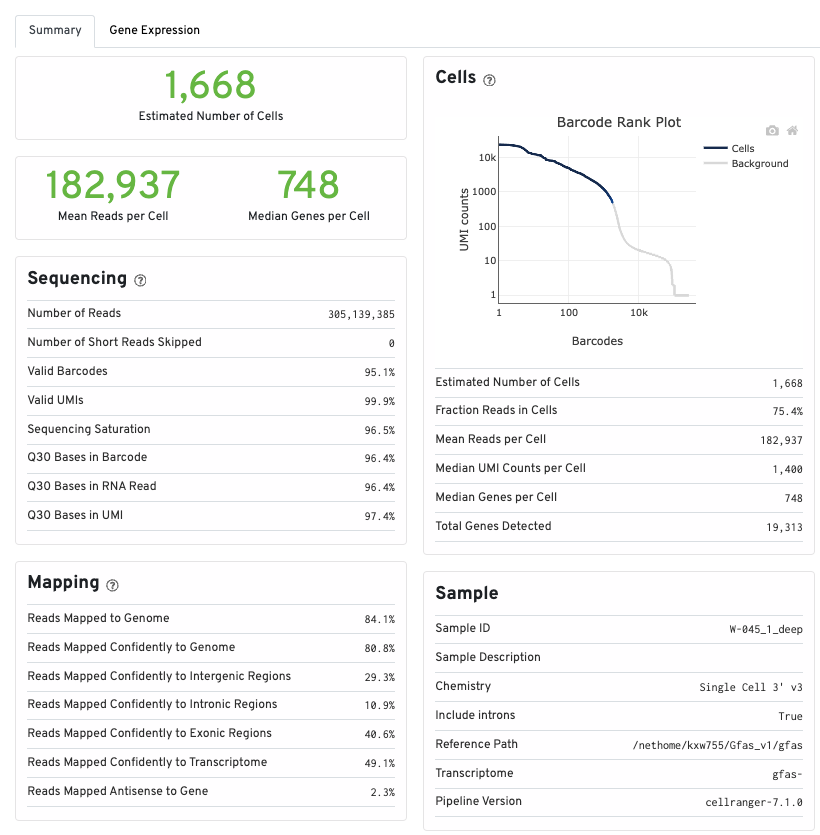

20231012
As per Levy et al 2021, they used the --force-cells flag to their estimated number of cells captured. I am going to try this with 5000 cells and see if the output differs.
Count
$ cd /nethome/kxw755/20231004_SingleCell_DG
$ nano count_W045_deep_force.job
#BSUB -J count_W045_deep_force
#BSUB -q general
#BSUB -P dark_genes
#BSUB -n 6
#BSUB -W 120:00
#BSUB -u kxw755@earth.miami.edu
#BSUB -o count.out
#BSUB -e count.err
#BSUB -B
#BSUB -N
###################################################################
cellranger count \
--id=W-045_1_deep_force \
--transcriptome=/nethome/kxw755/Gfas_v1/gfas \
--fastqs=/nethome/kxw755/20231004_SingleCell_DG \
--sample=AndradeRodriguez-15275-001_GEX3 \
--force-cells=5000
$ bsub < count_W045_deep_force.job
Exporting files
scp -r kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/20231004_SingleCell_DG/W-045_1_deep_force/outs/filtered_feature_bc_matrix.h5 /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/W-045_1_deep_force_raw_feature_bc_matrix.h5
scp -r kxw755@pegasus.ccs.miami.edu:/nethome/kxw755/20231004_SingleCell_DG/W-045_1_deep_force/outs/web_summary.html /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/W-045_1_deep_force_web_summary.html
Summary


20231128 - Analysis with new sequences
Today we recieved 10X Chromium data back from three bleached samples:
- S1 = Bleached (menthol)
- S2 = Bleached (menthol)
- S3 = Bleached (temperature)
The sequencing facility did mention that the runs were unbalanced, therefore we may need to resequence. This is also apparent because the file sizes greatly differ between libraries.
Download data from basespace and upload to Pegasus
cnidarianimmunity@Cnidarians-Mac-mini 16000-001_GEX3_S1 % md5 *gz
MD5 (AndradeRodriguez-16000-001_GEX3_S1_L003_R1_001.fastq.gz) = 0509ab579a8302d2dad5d6005ba7a53f
MD5 (AndradeRodriguez-16000-001_GEX3_S1_L003_R2_001.fastq.gz) = 5229e27eb1253012ac2302fedd286402
MD5 (AndradeRodriguez-16000-001_GEX3_S1_L005_R1_001.fastq.gz) = be95fd8c9c2e35e92e233d0499b2d9b8
MD5 (AndradeRodriguez-16000-001_GEX3_S1_L005_R2_001.fastq.gz) = 596678cb8f08de73f7e15b356e96b9ea
MD5 (AndradeRodriguez-16000-001_GEX3_S1_L006_R1_001.fastq.gz) = 45b7c0cf2416d81874d81184a350e0ac
MD5 (AndradeRodriguez-16000-001_GEX3_S1_L006_R2_001.fastq.gz) = 07118cc601cabb199b17862f2565b7e8
cnidarianimmunity@Cnidarians-Mac-mini 16000-002_GEX3_S2 % md5 *gz
MD5 (AndradeRodriguez-16000-002_GEX3_S2_L003_R1_001.fastq.gz) = 8984816bbda5a211cf33407d1cc3ee52
MD5 (AndradeRodriguez-16000-002_GEX3_S2_L003_R2_001.fastq.gz) = 8c9fe770c68995a6f3a2b95c709a0e63
MD5 (AndradeRodriguez-16000-002_GEX3_S2_L005_R1_001.fastq.gz) = 20b78b32cb2e82a219bc8a3e07e729fd
MD5 (AndradeRodriguez-16000-002_GEX3_S2_L005_R2_001.fastq.gz) = 17ddf534e38d9eaf1c400542d714ac04
MD5 (AndradeRodriguez-16000-002_GEX3_S2_L006_R1_001.fastq.gz) = 3f40c16136672822205bd710406df0ba
MD5 (AndradeRodriguez-16000-002_GEX3_S2_L006_R2_001.fastq.gz) = b2de3bb63694119a2b4f3d43b5483a48
cnidarianimmunity@Cnidarians-Mac-mini 16000-003_GEX3_S3 % md5 *gz
MD5 (AndradeRodriguez-16000-003_GEX3_S3_L003_R1_001.fastq.gz) = 1b8cd4810fa19725a4a385c73a73a15f
MD5 (AndradeRodriguez-16000-003_GEX3_S3_L003_R2_001.fastq.gz) = 8560db1974a914d5d1ef7fb3441b8456
MD5 (AndradeRodriguez-16000-003_GEX3_S3_L005_R1_001.fastq.gz) = 6d0c9d594fac387a3a3268b7bdcbe758
MD5 (AndradeRodriguez-16000-003_GEX3_S3_L005_R2_001.fastq.gz) = d8b81b64bb1ee970c428a12ef65aabf8
MD5 (AndradeRodriguez-16000-003_GEX3_S3_L006_R1_001.fastq.gz) = edaec25952b9ee5428b791087b41a90b
MD5 (AndradeRodriguez-16000-003_GEX3_S3_L006_R2_001.fastq.gz) = 825745ba7fb6e01874743ec91b7c5466
scp -r 20231128_BCL_Convert_FASTQ kxw755@pegasus.ccs.miami.edu:/scratch/projects/dark_genes/
md5 on samples after Pegasus transfer
md5sum *.gz
0509ab579a8302d2dad5d6005ba7a53f AndradeRodriguez-16000-001_GEX3_S1_L003_R1_001.fastq.gz
5229e27eb1253012ac2302fedd286402 AndradeRodriguez-16000-001_GEX3_S1_L003_R2_001.fastq.gz
be95fd8c9c2e35e92e233d0499b2d9b8 AndradeRodriguez-16000-001_GEX3_S1_L005_R1_001.fastq.gz
596678cb8f08de73f7e15b356e96b9ea AndradeRodriguez-16000-001_GEX3_S1_L005_R2_001.fastq.gz
45b7c0cf2416d81874d81184a350e0ac AndradeRodriguez-16000-001_GEX3_S1_L006_R1_001.fastq.gz
07118cc601cabb199b17862f2565b7e8 AndradeRodriguez-16000-001_GEX3_S1_L006_R2_001.fastq.gz
8984816bbda5a211cf33407d1cc3ee52 AndradeRodriguez-16000-002_GEX3_S2_L003_R1_001.fastq.gz
8c9fe770c68995a6f3a2b95c709a0e63 AndradeRodriguez-16000-002_GEX3_S2_L003_R2_001.fastq.gz
20b78b32cb2e82a219bc8a3e07e729fd AndradeRodriguez-16000-002_GEX3_S2_L005_R1_001.fastq.gz
17ddf534e38d9eaf1c400542d714ac04 AndradeRodriguez-16000-002_GEX3_S2_L005_R2_001.fastq.gz
3f40c16136672822205bd710406df0ba AndradeRodriguez-16000-002_GEX3_S2_L006_R1_001.fastq.gz
b2de3bb63694119a2b4f3d43b5483a48 AndradeRodriguez-16000-002_GEX3_S2_L006_R2_001.fastq.gz
1b8cd4810fa19725a4a385c73a73a15f AndradeRodriguez-16000-003_GEX3_S3_L003_R1_001.fastq.gz
8560db1974a914d5d1ef7fb3441b8456 AndradeRodriguez-16000-003_GEX3_S3_L003_R2_001.fastq.gz
6d0c9d594fac387a3a3268b7bdcbe758 AndradeRodriguez-16000-003_GEX3_S3_L005_R1_001.fastq.gz
d8b81b64bb1ee970c428a12ef65aabf8 AndradeRodriguez-16000-003_GEX3_S3_L005_R2_001.fastq.gz
edaec25952b9ee5428b791087b41a90b AndradeRodriguez-16000-003_GEX3_S3_L006_R1_001.fastq.gz
825745ba7fb6e01874743ec91b7c5466 AndradeRodriguez-16000-003_GEX3_S3_L006_R2_001.fastq.gz
Sample 1 Count
cd /scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-001_GEX3_S1
nano count_S1.job
#BSUB -J count_S1
#BSUB -q general
#BSUB -P dark_genes
#BSUB -n 6
#BSUB -W 120:00
#BSUB -u kxw755@earth.miami.edu
#BSUB -o count.out
#BSUB -e count.err
#BSUB -B
#BSUB -N
###################################################################
cellranger count \
--id=S1 \
--transcriptome=/nethome/kxw755/Gfas_v1/gfas \
--fastqs=/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-001_GEX3_S1 \
--sample=AndradeRodriguez-16000-001_GEX3
` bsub < count_S1.job`
scp -r kxw755@pegasus.ccs.miami.edu:/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-001_GEX3_S1/S1/outs/filtered_feature_bc_matrix.h5 /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/S1_raw_feature_bc_matrix.h5
scp -r kxw755@pegasus.ccs.miami.edu:/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-001_GEX3_S1/S1/outs/web_summary.html /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/S1_web_summary.html
scp -r kxw755@pegasus.ccs.miami.edu:/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-001_GEX3_S1/S1/outs/metrics_summary.csv /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/S1_metrics_summary.csv
## Sample 2 Count
cd /scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-002_GEX3_S2
nano count_S2.job
#BSUB -J count_S2
#BSUB -q general
#BSUB -P dark_genes
#BSUB -n 6
#BSUB -W 120:00
#BSUB -u kxw755@earth.miami.edu
#BSUB -o count.out
#BSUB -e count.err
#BSUB -B
#BSUB -N
###################################################################
cellranger count \
--id=S2 \
--transcriptome=/nethome/kxw755/Gfas_v1/gfas \
--fastqs=/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-002_GEX3_S2 \
--sample=AndradeRodriguez-16000-002_GEX3
bsub < count_S2.job
scp -r kxw755@pegasus.ccs.miami.edu:/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-002_GEX3_S2/S2/outs/filtered_feature_bc_matrix.h5 /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/S2_raw_feature_bc_matrix.h5
scp -r kxw755@pegasus.ccs.miami.edu:/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-002_GEX3_S2/S2/outs/web_summary.html /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/S2_web_summary.html
scp -r kxw755@pegasus.ccs.miami.edu:/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-002_GEX3_S2/S2/outs/metrics_summary.csv /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/S2_metrics_summary.csv
Sample 3 Count
cd /scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-003_GEX3_S3
nano count_S3.job
#BSUB -J count_S3
#BSUB -q general
#BSUB -P dark_genes
#BSUB -n 6
#BSUB -W 120:00
#BSUB -u kxw755@earth.miami.edu
#BSUB -o count.out
#BSUB -e count.err
#BSUB -B
#BSUB -N
###################################################################
cellranger count \
--id=S3 \
--transcriptome=/nethome/kxw755/Gfas_v1/gfas \
--fastqs=/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-003_GEX3_S3 \
--sample=AndradeRodriguez-16000-003_GEX3
bsub < count_S3.job
scp -r kxw755@pegasus.ccs.miami.edu:/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-003_GEX3_S3/S3/outs/filtered_feature_bc_matrix.h5 /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/S3_raw_feature_bc_matrix.h5
scp -r kxw755@pegasus.ccs.miami.edu:/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-003_GEX3_S3/S3/outs/web_summary.html /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/S3_web_summary.html
scp -r kxw755@pegasus.ccs.miami.edu:/scratch/projects/dark_genes/20231128_BCL_Convert_FASTQ/16000-003_GEX3_S3/S3/outs/metrics_summary.csv /Users/kevinwong/MyProjects/DarkGenes_Bleaching_Comparison/output/CellRanger/S3_metrics_summary.csv

