Methylseq trimming test to remove m-bias
Goal
- To remove the M-bias at the beginning of R2 for some WBGS samples. This seems to be a common problem for WGBS using library preps like Zymo.
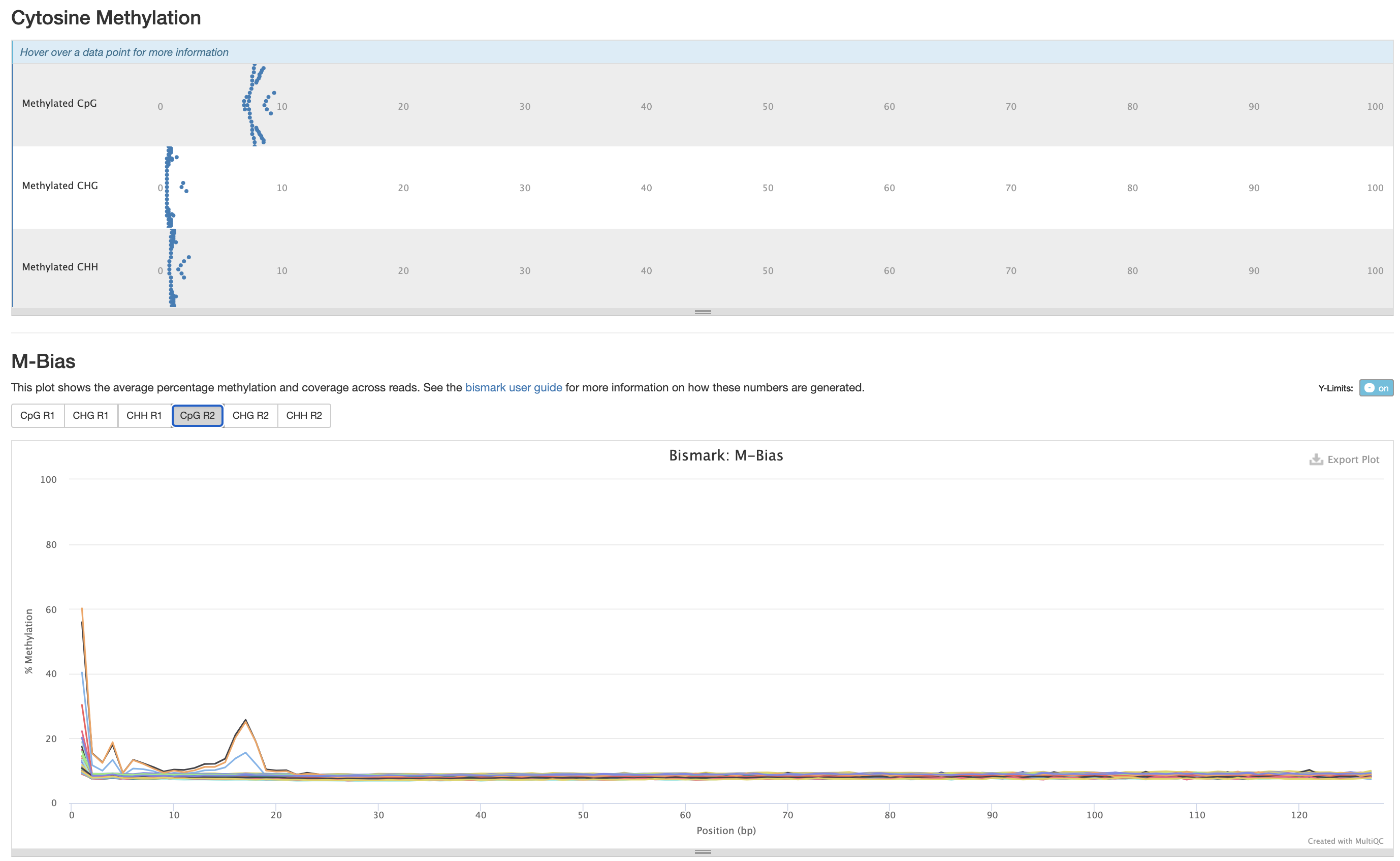
Trimming Parameter testing
I am testing the following parameters below to see which trimming parameters remove the m-bias in one test sample (18-9):
| Clipping Parameters | R1 5’ | R2 5’ | R1’3 | R2 3’ |
|---|---|---|---|---|
| Original | 10 | 10 | 10 | 10 |
| Zymo Preset | 10 | 15 | 10 | 10 |
| Trim_1 | 10 | 30 | 10 | 10 |
| Trim_2 | 10 | 30 | 30 | 10 |
| Trim_3 | 15 | 30 | 30 | 15 |
| Trim_4 | 15 | 30 | 15 | 15 |
Results
All tests were run on the same sample file: 18-9_S159_L004_R1_001_val_1
Summary Table
| Original | Zymo Preset | Trim_1 | Trim_2 | Trim_3 | Trim_4 | |
|---|---|---|---|---|---|---|
| % mCpG | 9.40% | 9.20% | 8.90% | 8.90% | 8.50% | 8.50% |
| % mCHG | 2.20% | 2.10% | 1.90% | 1.80% | 1.60% | 1.60% |
| % mCHH | 2.40% | 2.40% | 2.20% | 2.40% | 2.10% | 2.40% |
| M C’s | 141.5 | 144.1 | 146.1 | 125.7 | 133.9 | 119.4 |
| % Dups | 11.90% | 11.90% | 11.80% | 11.80% | 11.80% | 11.80% |
| % Aligned | 26.10% | 27.60% | 31.20% | 30.40% | 32.50% | 31.60% |
| Ins. size | 103 | 102 | 99 | 89 | 93 | 86 |
| Median cov | 0.0X | 0.0X | 0.0X | 0.0X | 0.0X | 0.0X |
| Mean cov | 1.9X | 1.9X | 1.8X | 1.6X | 1.7X | 1.5X |
| R1_% BP Trimmed | 46.90% | 46.90% | 46.90% | 46.90% | 46.90% | 46.90% |
| R1_% Dups | 15.70% | 15.70% | 15.70% | 15.70% | 15.70% | 15.70% |
| R1_% GC | 42% | 42% | 42% | 42% | 42% | 42% |
| R1_M Seqs | 47.7 | 47.7 | 47.7 | 47.7 | 47.7 | 47.7 |
| R2_% BP Trimmed | 42.70% | 42.70% | 42.70% | 42.70% | 43% | 42.70% |
| R2_% Dups | 16.80% | 16.80% | 16.80% | 16.80% | 16.80% | 16.80% |
| R2_% GC | 41% | 41% | 41% | 41.00% | 41% | 41.00% |
| R2_M Seqs | 47.7 | 47.7 | 47.7 | 47.7 | 47.7 | 47.7 |
MultiQC reports
Investigating m-bias plots on R2
Zymo Preset
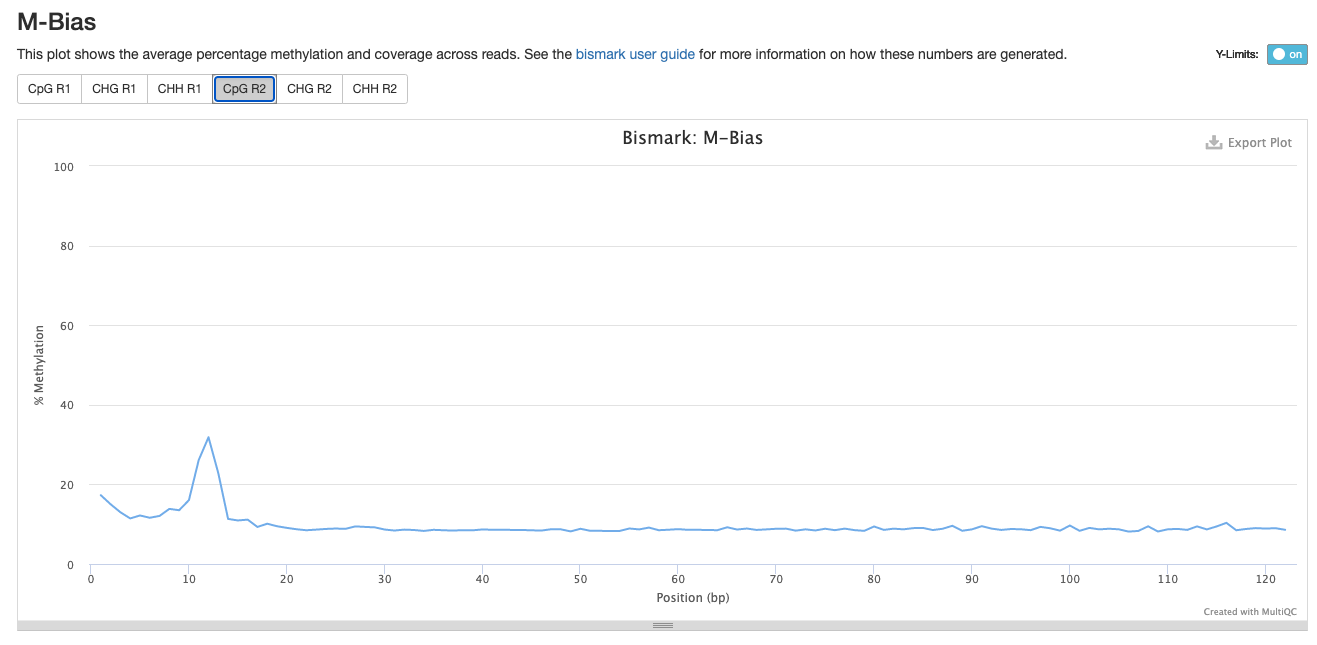
Trim_1
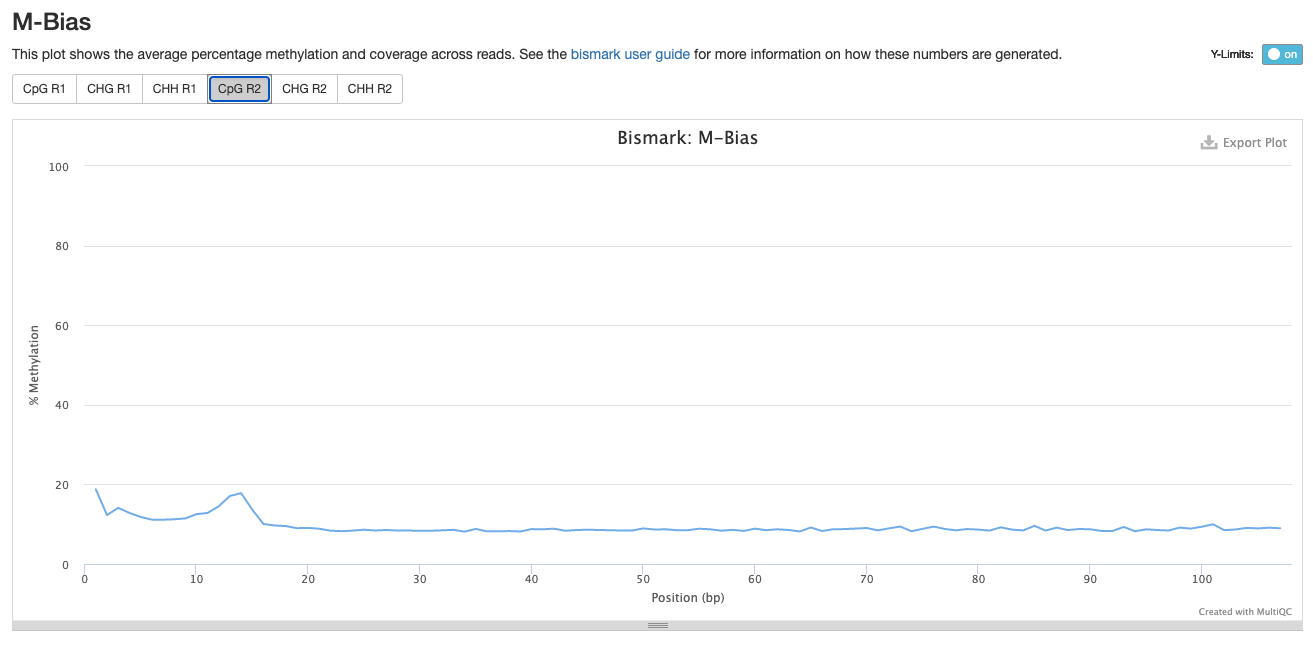
Trim_2
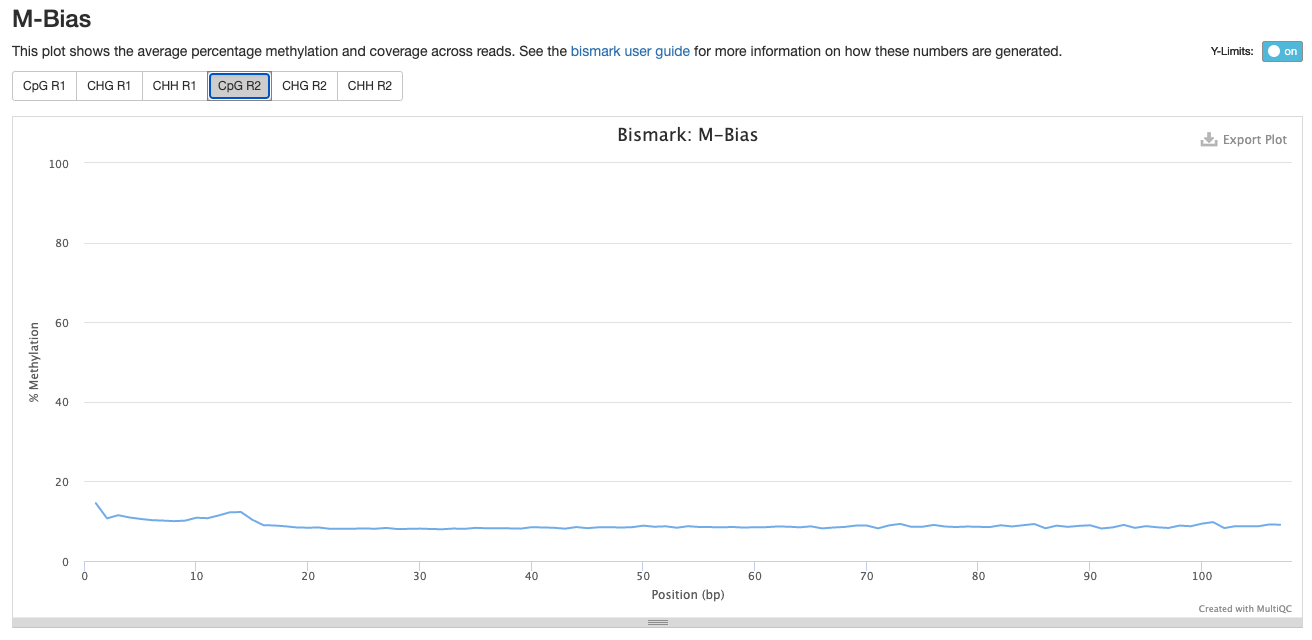
Trim_3
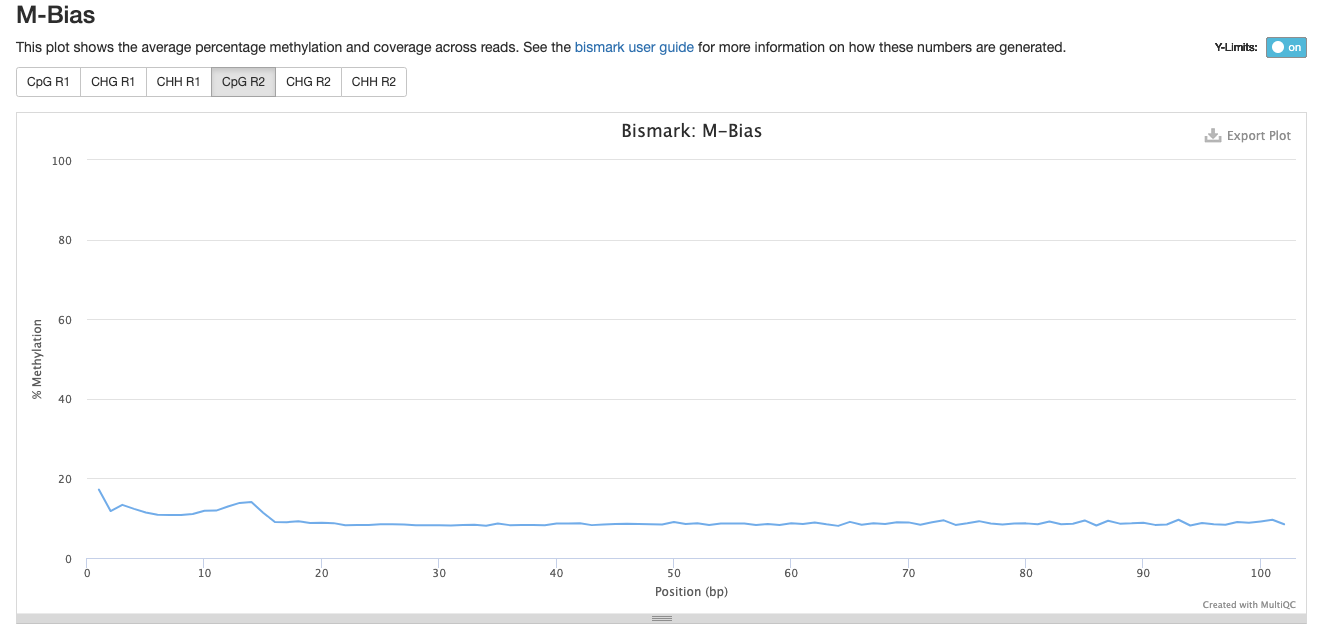
Trim_4
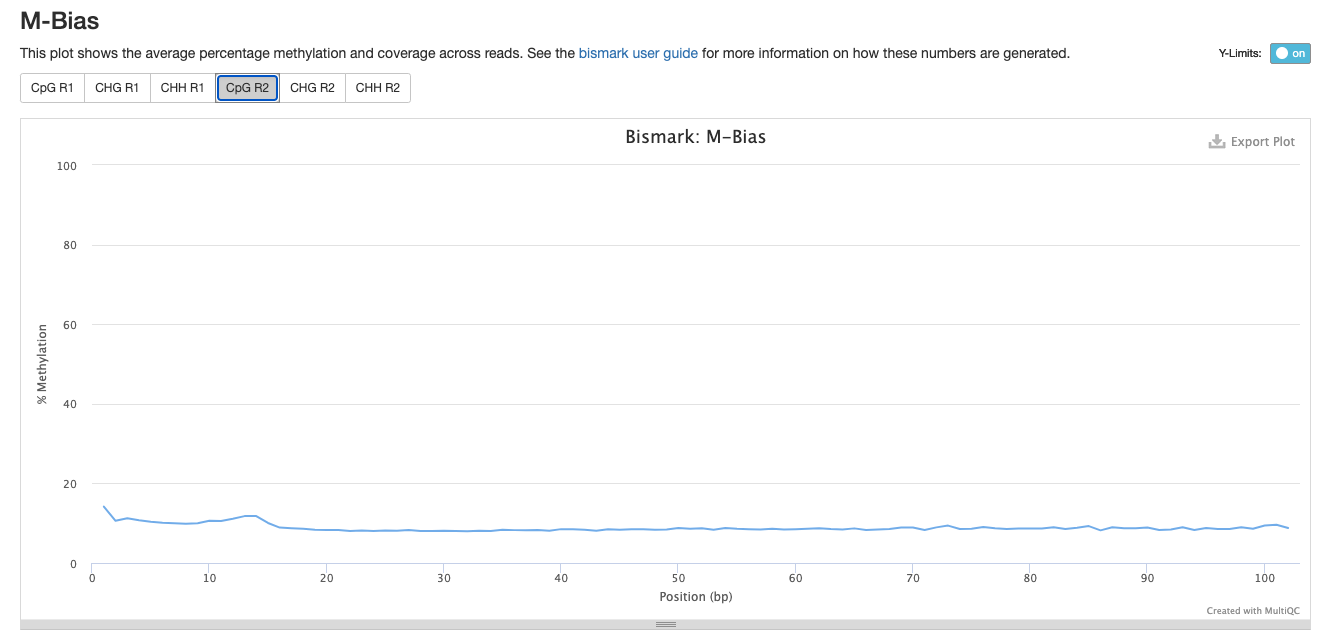
Investigating m-bias plots on R1
For Trim_3 and Trim_4 Parameters, the % methylation also decreases at the beginning of the R1:
Trim_2 R1
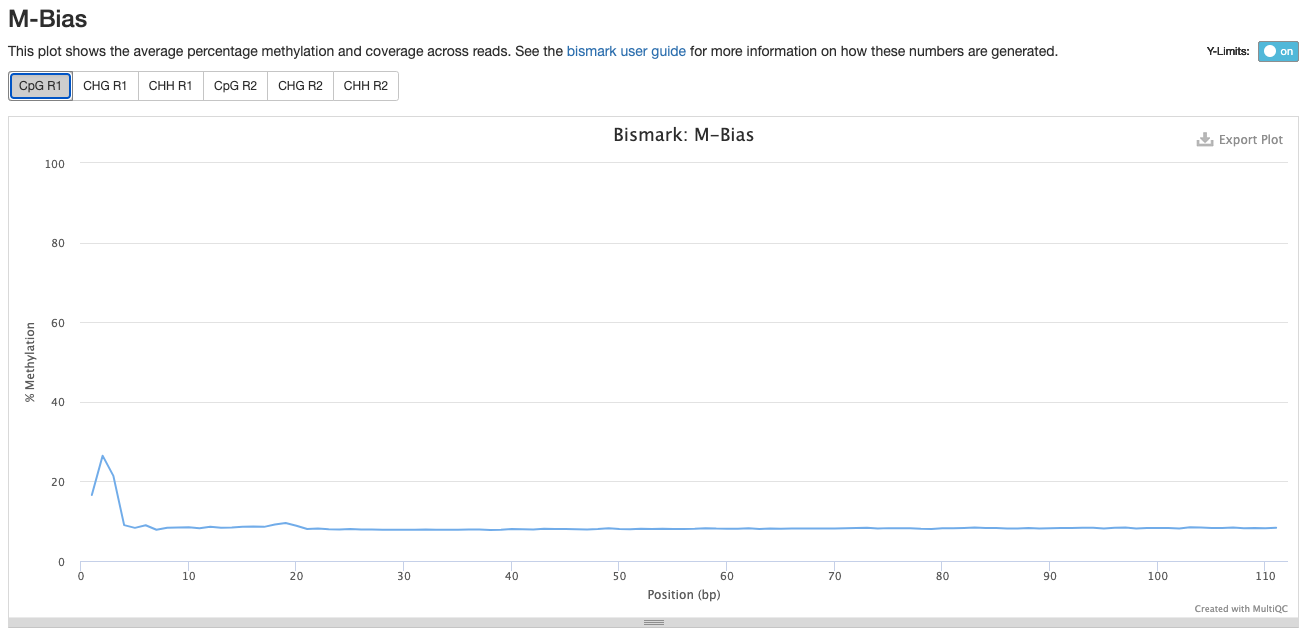
Trim_3 R1
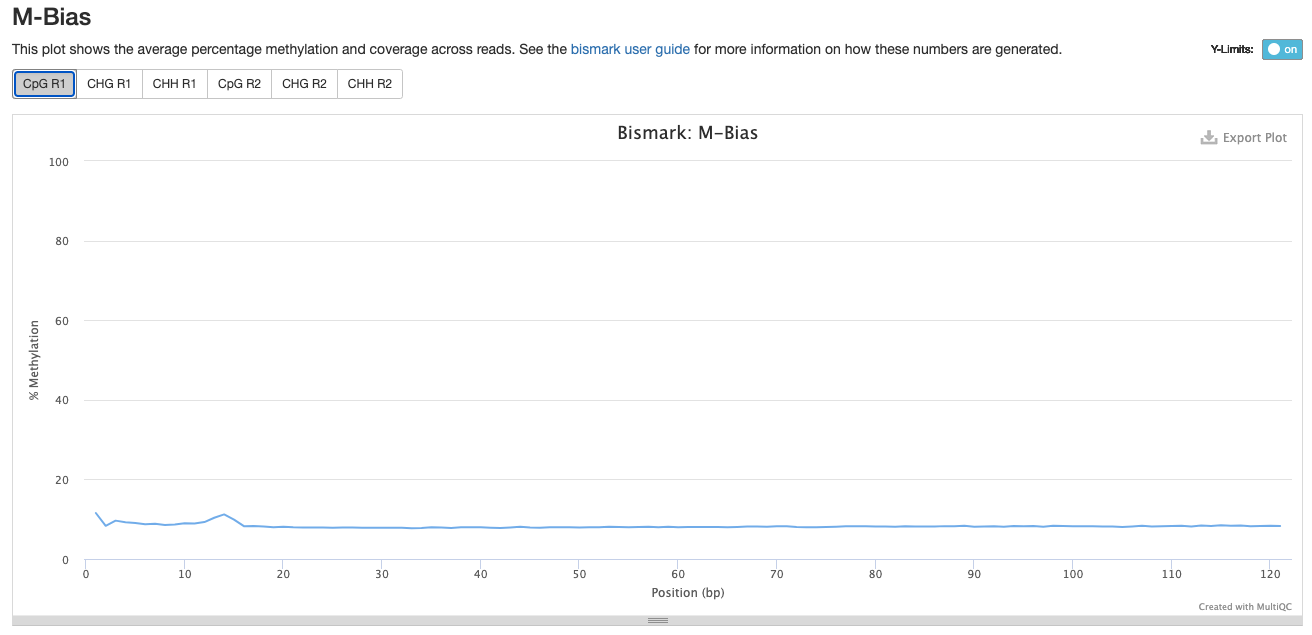
Scripts
Zymo presets
#!/bin/bash
#SBATCH --job-name="methylseq"
#SBATCH -t 120:00:00
#SBATCH --mem=120GB
#SBATCH --nodes=1 --ntasks-per-node=10
#SBATCH --account=putnamlab
#SBATCH --export=NONE
#SBATCH --mail-type=BEGIN,END,FAIL
#SBATCH --mail-user=kevin_wong1@uri.edu
#SBATCH -D /data/putnamlab/kevin_wong1/Thermal_Transplant_WGBS/test_zymo
#SBATCH --exclusive
#SBATCH --cpus-per-task=3
# load modules needed
module load Nextflow/21.03.0
# run nextflow methylseq
nextflow run nf-core/methylseq \
-profile singularity \
--aligner bismark \
--igenomes_ignore \
--fasta /data/putnamlab/kevin_wong1/Past_Genome/past_filtered_assembly.fasta \
--save_reference \
--input '/data/putnamlab/KITT/hputnam/20211008_Past_ThermalTransplant_WGBS/18-9_S159_L004_R{1,2}_001.fastq.gz' \
--zymo \
--non_directional \
--cytosine_report \
--relax_mismatches \
--unmapped \
--outdir WGBS_methylseq /
-name WGBS_methylseq_past_zymo
scp kevin_wong1@ssh3.hac.uri.edu:/data/putnamlab/kevin_wong1/Thermal_Transplant_WGBS/test_zymo/WGBS_methylseq/MultiQC/multiqc_report.html MyProjects/Thermal_Transplant_Molecular/output/multiqc_report_zymotest.html
Trim_1
#!/bin/bash
#SBATCH --job-name="methylseq"
#SBATCH -t 120:00:00
#SBATCH --nodes=1 --ntasks-per-node=10
#SBATCH --mem=500GB
#SBATCH --account=putnamlab
#SBATCH --export=NONE
#SBATCH --mail-type=BEGIN,END,FAIL
#SBATCH --mail-user=kevin_wong1@uri.edu
#SBATCH -p putnamlab
#SBATCH -D /data/putnamlab/kevin_wong1/Thermal_Transplant_WGBS/test_trim
#SBATCH --exclusive
#SBATCH --cpus-per-task=3
# load modules needed
module load Nextflow/21.03.0
# run nextflow methylseq
nextflow run nf-core/methylseq \
-profile singularity \
--aligner bismark \
--igenomes_ignore \
--fasta /data/putnamlab/kevin_wong1/Past_Genome/past_filtered_assembly.fasta \
--save_reference \
--input '/data/putnamlab/KITT/hputnam/20211008_Past_ThermalTransplant_WGBS/18-9_S159_L004_R{1,2}_001.fastq.gz' \
--clip_r1 10 \
--clip_r2 30 \
--three_prime_clip_r1 10 --three_prime_clip_r2 10 \
--non_directional \
--cytosine_report \
--relax_mismatches \
--unmapped \
--outdir WGBS_methylseq
-name WGBS_methylseq_past_trim
scp kevin_wong1@ssh3.hac.uri.edu:/data/putnamlab/kevin_wong1/Thermal_Transplant_WGBS/test_trim/WGBS_methylseq/MultiQC/multiqc_report.html MyProjects/Thermal_Transplant_Molecular/output/multiqc_report_trimtest.html
Trim_2
#!/bin/bash
#SBATCH --job-name="methylseq"
#SBATCH -t 120:00:00
#SBATCH --nodes=1 --ntasks-per-node=10
#SBATCH --mem=120GB
#SBATCH --account=putnamlab
#SBATCH --export=NONE
#SBATCH --mail-type=BEGIN,END,FAIL
#SBATCH --mail-user=kevin_wong1@uri.edu
#SBATCH -D /data/putnamlab/kevin_wong1/Thermal_Transplant_WGBS/test_trim_2
#SBATCH --exclusive
# load modules needed
module load Nextflow/21.03.0
# run nextflow methylseq
nextflow run nf-core/methylseq \
-profile singularity \
--aligner bismark \
--igenomes_ignore \
--fasta /data/putnamlab/kevin_wong1/Past_Genome/past_filtered_assembly.fasta \
--save_reference \
--input '/data/putnamlab/KITT/hputnam/20211008_Past_ThermalTransplant_WGBS/18-9_S159_L004_R{1,2}_001.fastq.gz' \
--clip_r1 10 \
--clip_r2 30 \
--three_prime_clip_r1 30 \
--three_prime_clip_r2 10 \
--non_directional \
--cytosine_report \
--relax_mismatches \
--unmapped \
--outdir WGBS_methylseq
scp kevin_wong1@ssh3.hac.uri.edu:/data/putnamlab/kevin_wong1/Thermal_Transplant_WGBS/test_trim_2/WGBS_methylseq/MultiQC/multiqc_report.html MyProjects/Thermal_Transplant_Molecular/output/multiqc_report_trimtest2.html
Trim_3
#!/bin/bash
#SBATCH --job-name="methylseq"
#SBATCH -t 120:00:00
#SBATCH --nodes=1 --ntasks-per-node=10
#SBATCH --mem=120GB
#SBATCH --account=putnamlab
#SBATCH --export=NONE
#SBATCH --mail-type=BEGIN,END,FAIL
#SBATCH --mail-user=kevin_wong1@uri.edu
#SBATCH -D /data/putnamlab/kevin_wong1/Thermal_Transplant_WGBS/test_trim_3
#SBATCH --exclusive
# load modules needed
module load Nextflow/21.03.0
# run nextflow methylseq
nextflow run nf-core/methylseq \
-profile singularity \
--aligner bismark \
--igenomes_ignore \
--fasta /data/putnamlab/kevin_wong1/Past_Genome/past_filtered_assembly.fasta \
--save_reference \
--input '/data/putnamlab/KITT/hputnam/20211008_Past_ThermalTransplant_WGBS/18-9_S159_L004_R{1,2}_001.fastq.gz' \
--clip_r1 15 \
--clip_r2 30 \
--three_prime_clip_r1 30 \
--three_prime_clip_r2 15 \
--non_directional \
--cytosine_report \
--relax_mismatches \
--unmapped \
--outdir WGBS_methylseq
scp kevin_wong1@ssh3.hac.uri.edu:/data/putnamlab/kevin_wong1/Thermal_Transplant_WGBS/test_trim_3/WGBS_methylseq/MultiQC/multiqc_report.html MyProjects/Thermal_Transplant_Molecular/output/multiqc_report_trimtest3.html
Trim_4
#!/bin/bash
#SBATCH --job-name="methylseq"
#SBATCH -t 120:00:00
#SBATCH --nodes=1 --ntasks-per-node=10
#SBATCH --mem=120GB
#SBATCH --account=putnamlab
#SBATCH --export=NONE
#SBATCH --mail-type=BEGIN,END,FAIL
#SBATCH --mail-user=kevin_wong1@uri.edu
#SBATCH -D /data/putnamlab/kevin_wong1/Thermal_Transplant_WGBS/test_trim_4
#SBATCH --exclusive
# load modules needed
module load Nextflow/21.03.0
# run nextflow methylseq
nextflow run nf-core/methylseq \
-profile singularity \
--aligner bismark \
--igenomes_ignore \
--fasta /data/putnamlab/kevin_wong1/Past_Genome/past_filtered_assembly.fasta \
--save_reference \
--input '/data/putnamlab/KITT/hputnam/20211008_Past_ThermalTransplant_WGBS/18-9_S159_L004_R{1,2}_001.fastq.gz' \
--clip_r1 15 \
--clip_r2 30 \
--three_prime_clip_r1 15 \
--three_prime_clip_r2 15 \
--non_directional \
--cytosine_report \
--relax_mismatches \
--unmapped \
--outdir WGBS_methylseq
scp kevin_wong1@ssh3.hac.uri.edu:/data/putnamlab/kevin_wong1/Thermal_Transplant_WGBS/test_trim_4/WGBS_methylseq/MultiQC/multiqc_report.html MyProjects/Thermal_Transplant_Molecular/output/multiqc_report_trimtest4.html
Written on November 16, 2021
