Zymo-DNA-RNA-Extract-P.astreoides-Genome
RNA/DNA extractions for P. astreoides genome samples
Kit: ZYMO Quick-DNA/RNA Miniprep Plus Kit
Goal
- To extract DNA and RNA for an Illumina RNAseq library prep workshop
Experimental design
Table 1: Sample descriptions for this extraction
| Sample # | Sample Type | Coral ID | Size of Sample | Treatment |
|---|---|---|---|---|
| 1 | Fragment | PG #1 | ~10mm diameter | 23°C + Air |
| 2 | Fragment | PG #2 | ~10mm diameter | -22°C + Dark |
- I sampled duplicates of each sample, preserved them in 500μL DNA/RNA shield in ZR BBashing lysis tubes with 0.1mm and 0.5mm beads and stored at -80°C
Overall workflow
- Prepare samples
- Extract and prep DNA with column (elute RNA and Protein)
- Extract and prep RNA with column (elute Protein)
- Quantify DNA, RNA, and protein extracts
Protocol preparation
- Using slightly modified Zymo Duet DNA/RNA extraction protocol which will extract both DNA and RNA at the same time (Below are summary steps)
Reagents and supplies
- RNase-free Water
- 100% ethanol, ACS grade or better
- 10mM Tris HCl pH 8.0 made with RNase-free water
- ZR BBashing lysis tubes with 0.1mm and 0.5mm beads
Equipment
- Rocking oven that can be set to 70°C
- RNase away and a designated RNase free space
- Tabletop and larger centrifuges for 1.5mL and 50mL tubes capable of 12,000 x g
Clipper Sterilization
- Rub down clippers with:
- 10% Bleach solution
- DI water
- 90% ethanol
- RNAse free water
Notes before starting
- Wipe down benchtop with RNase away and have the spray bottle and kimwipes on-hand to use frequently
Sample preparation
Fragment preparation
- Sterilize clippers (as outlined above)
- Clip off ~10mm in diameter of tissue and skeleton and place into ZR BBashing lysis tube
- Add 500 μL of DNA/RNA shield
- Vortex for ~1 minute
- Remove supernatant and transfer into 1.5mL tube
- I was able to remove ~450μL
DNA Extraction
- Add equal volumes of DNA lysis buffer as sample volume to the 5 mL tube with sample
- 450 μL for the fragment samples
- Vortex to mix
- Transfer 700 μL into DNA filter column (yellow)
- Centrifuge at 16,000 rcf for 30 seconds
- Remove flow through liquid and transfer into a new 5 mL tube labeled for RNA
- Repeat steps 2 until all liquid is gone
- Add 400 μL of DNA/RNA prep buffer to column
- Centrifuge at 16,000 rcf for 30 seconds
- Remove the flow through and transfer to waste
- Add 700 μL of DNA/RNA wash buffer
- Centrifuge at 16,000 rcf for 30 seconds
- Remove the flow through and transfer to waste
- Add 400 μL of DNA/RNA wash buffer
- Centrifuge at 16,000 rcf for 2 minutes
- Remove the flow through and transfer to waste
- Removed column and place into a new sterile 1.5 mL tube
- Add 50 μL of 10 mM Tris HCL (warmed to 55°C) directly to filter
- Incubate at room temperature for 5 minutes
- Centrifuge at 16,000 rcf for 30 seconds
- Keep flow through in tube
- Add another 50 μL of 10 mM Tris HCL (warmed to 55°C) directly to filter
- Incubate at room temperature for 5 minutes
- Centrifuge at 16,000 rcf for 30 seconds
- Discard column and keep flow through in tube
- After quantifying on the Qubit, I aliquoted into two 1.5 eppindorf tubes (10 μL and the remaining 89 μL) and stored at -20°C
- Tubes were labelled: DNA #1 KW 03/13 and DNA #2 KW 03/13
RNA Extraction
- Add equal volumes of 100% Ethanol as sample volume to the 5 mL tube with sample
- 900 μL for the fragment samples
- Vortex to mix
- Transfer 700 μL into RNA filter column (green)
- Centrifuge at 16,000 rcf for 30 seconds
- Remove flow through liquid and transfer into a new 5 mL tube labeled for Protein and store in -80°C
- Repeat steps 12 until all liquid is gone
- Add 400 μL of DNA/RNA wash buffer to column
- Centrifuge at 16,000 rcf for 30 seconds
- Remove the flow through and transfer to waste
- Make DNase I master mix: [75μL DNA digestion buffer and 5μL DNase] X n (sample #)
- Two samples, 150μL DNA digestion buffer and 10μL DNase
- Add 80μL of DNase I master mix directly to the filter of each column. Incubate at room temperature for ~15 minutes
- Add 400 μL of DNA/RNA prep buffer to column
- Centrifuge at 16,000 rcf for 30 seconds
- Remove the flow through and transfer to waste
- Add 700 μL of DNA/RNA wash buffer
- Centrifuge at 16,000 rcf for 30 seconds
- Remove the flow through and transfer to waste
- Add 400 μL of DNA/RNA wash buffer
- Centrifuge at 16,000 rcf for 2 minutes
- Remove the flow through and transfer to waste
- Removed column and place into a new sterile 1.5 mL tube
- Add 50 μL of RNAase-free water (warmed to 55°C) directly to filter
- Incubate at room temperature for 5 minutes
- Centrifuge at 16,000 rcf for 30 seconds
- Keep flow through in tube
- Add another 50 μL of RNAase-free water (warmed to 55°C) directly to filter
- Incubate at room temperature for 5 minutes
- Centrifuge at 16,000 rcf for 30 seconds
- Discard column and keep flow through in tube
- Aliquot into appropriate tubes for storage at -80°C
Quantification
Qubit
- For the Qubit readings, 10 μL of each standard were used and 1 μL of each sample were used.
Table 2: Qubit readings for the extracted DNA and RNA samples.
| Standard 1 | Standard 2 | #1 (ng/uL) | #2 (ng/uL) | |
|---|---|---|---|---|
| DNA | 199.37 | 21533.38 | 10.8 | 29.2 |
| RNA | 401.46 | 11418.56 | 23.6 | 34.2 |
Tapestation
- Only RNA samples were run on the tape station
Tapestation results
Sample #1:
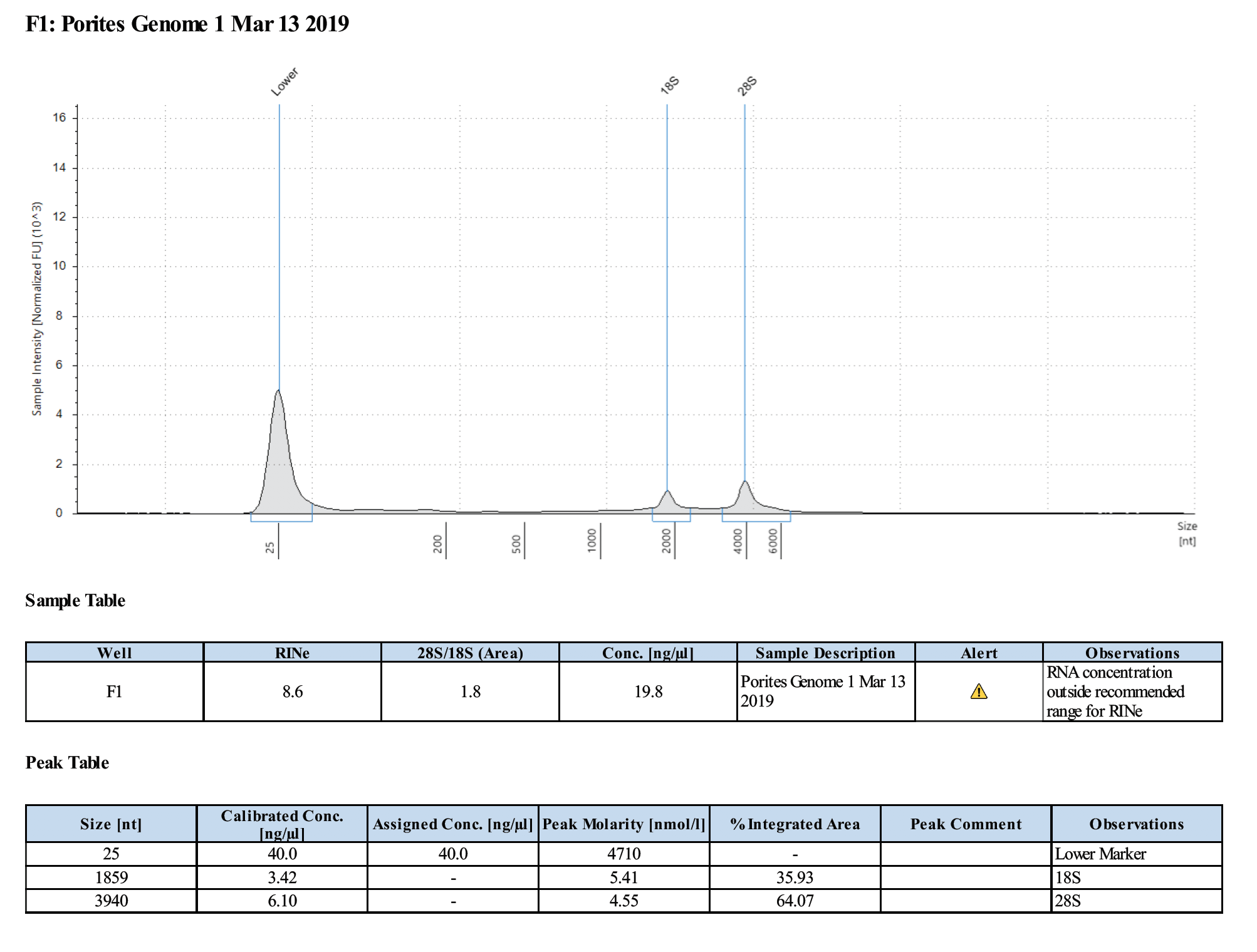
Sample #2:
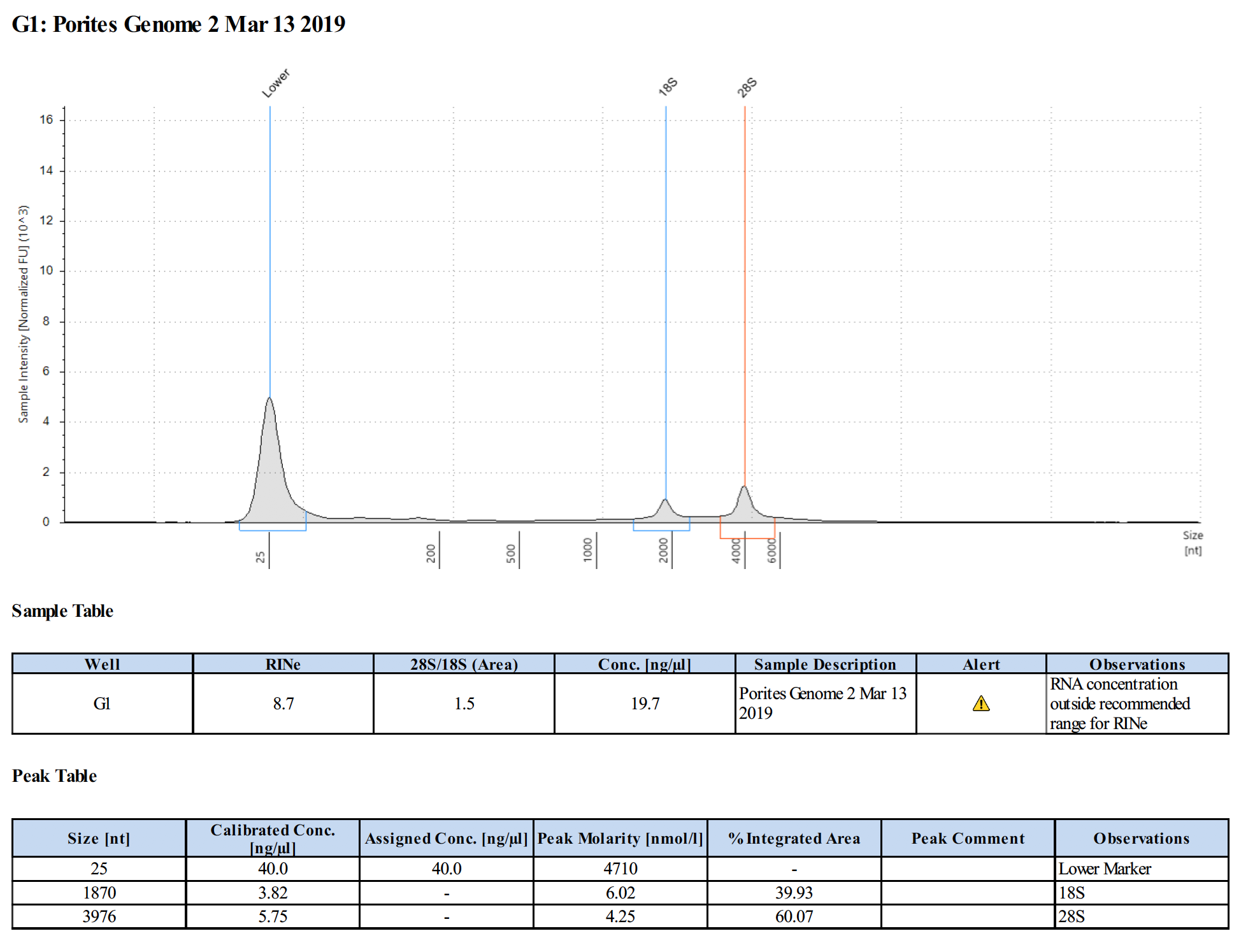
Conclusions
- Samples are good for the Illumina RNA-Seq workshop
- Changes in Ethanol addition in RNA extraction step potentially produced higher concentrations of RNA
Written on March 13, 2019
