20210422 WGBS PicoMethyl-seq library prep for Thermal Transplant Trial 3
Project: Thermal Transplant Molecular
Goal
Third trial of WGBS library prep using the Zymo Pico Methyl-seq library prep kit and this protocol.
Today, I wanted to test 2 different modifications during the second amplification:
Modification #1:
- Use the combined i5 and i7 index primers (an input of 1uL)
- Reduce the Library Amp Master Mix (2x) to 13uL (was originally 14uL)
Modification #2:
- Use the KAPA HiFi HotStart ReadyMix (2X) instead of the Library Amp Master Mix (2x)
Sample dilution and mix calculations can be found on this google sheet
Protocol
I am using a modified version of this protocol.
Samples and Indices
| Sample | Coral ID | Life Stage | i5 Index # | i7 Index # | Modification # |
|---|---|---|---|---|---|
| 18-118 | P-6-A | Adult | 7 | 7 | 1 |
| 18-106 | P-19-A | Adult | 8 | 8 | 1 |
| 18-334 | P-19-B | Adult | 9 | 9 | 2 |
| 18-442 | R-9-B | Adult | 10 | 10 | 2 |
| 18-167 | R-19-A | Adult | 11 | 11 | NA |
| L-1053 | P-12-A | Larvae | 12 | 12 | NA |
| L-1093 | R-19-A | Larvae | 13 | 13 | NA |
| L-704 | P-15-A | Larvae | 14 | 14 | NA |
Qubit Results
To test DNA quantity: Qubit
I used the DNA broad range assay.
I ran the qubit and tapestation on the following day (20210423). After the bead clean up step I stored the libraries in the fridge at 4C for ~16 hours.
| Sample | DNA 1 (ng/ul) | DNA 2 (ng/ul) |
|---|---|---|
| Standard 1 | 176.98 | |
| Standard 2 | 19057.22 | |
| 18-118 (#7) | 25.2 | 24.8 |
| 18-106 (#8) | 25.2 | 24.8 |
| 18-334 (#9) | 28.0 | 27.8 |
| 18-442 (#10) | 73.2 | 72.0 |
| 18-167 (#11) | 29.4 | 28.8 |
| L-1053 (#12) | 8.90 | 8.50 |
| L-1093 (#13) | 24.6 | 24.0 |
| L-704 (#14) | 7.08 | 6.92 |
Tapestation Results
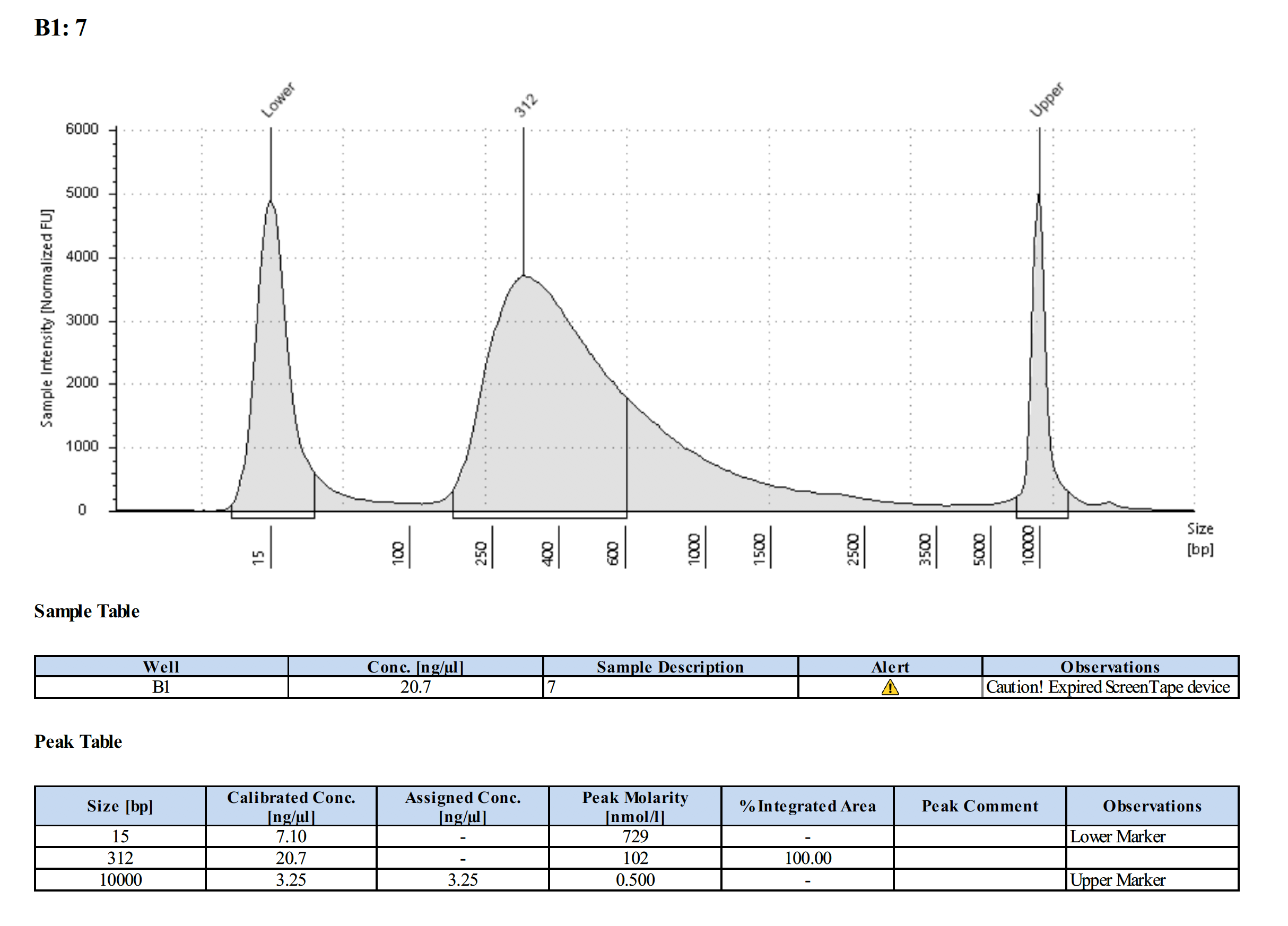
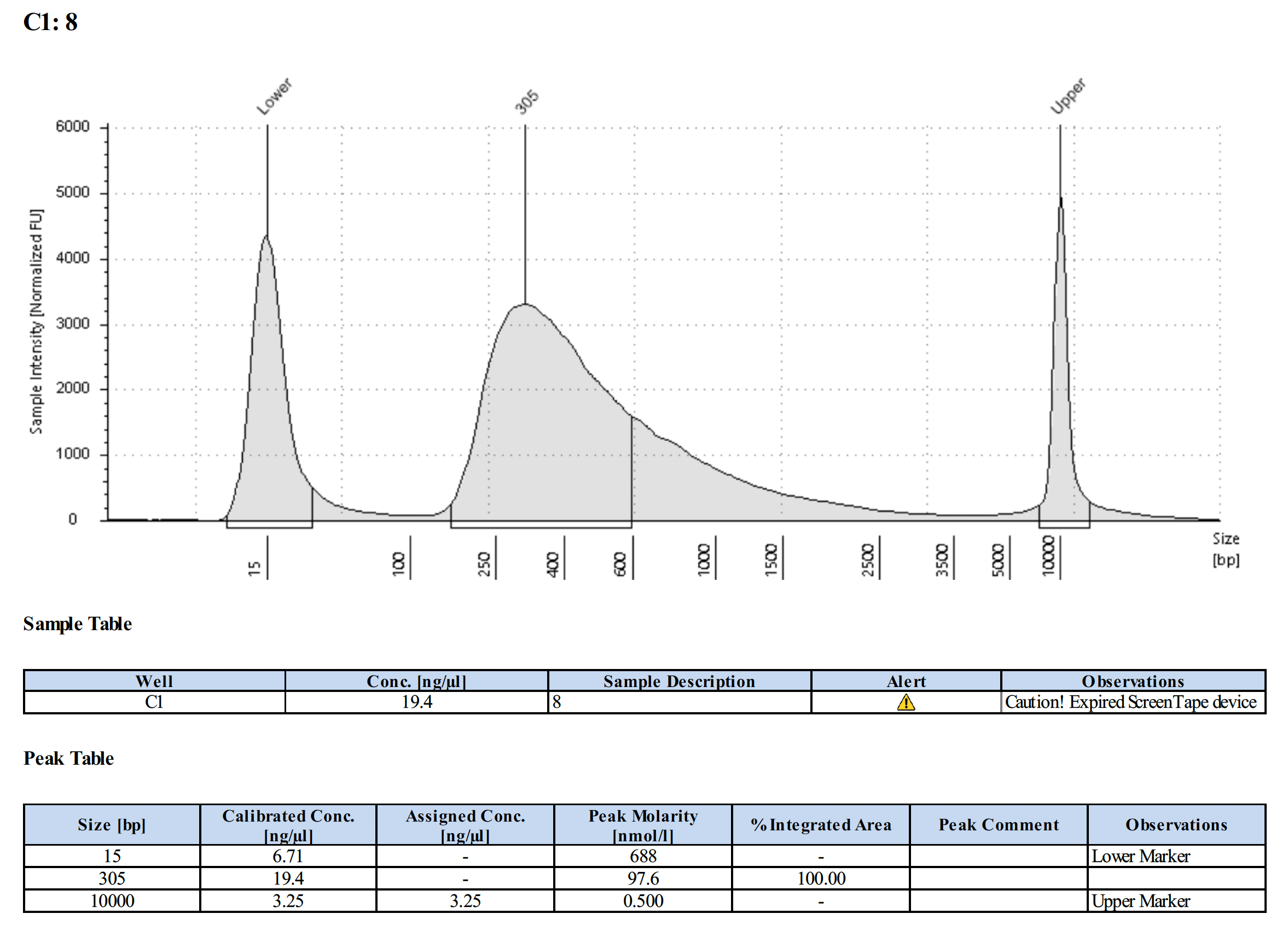
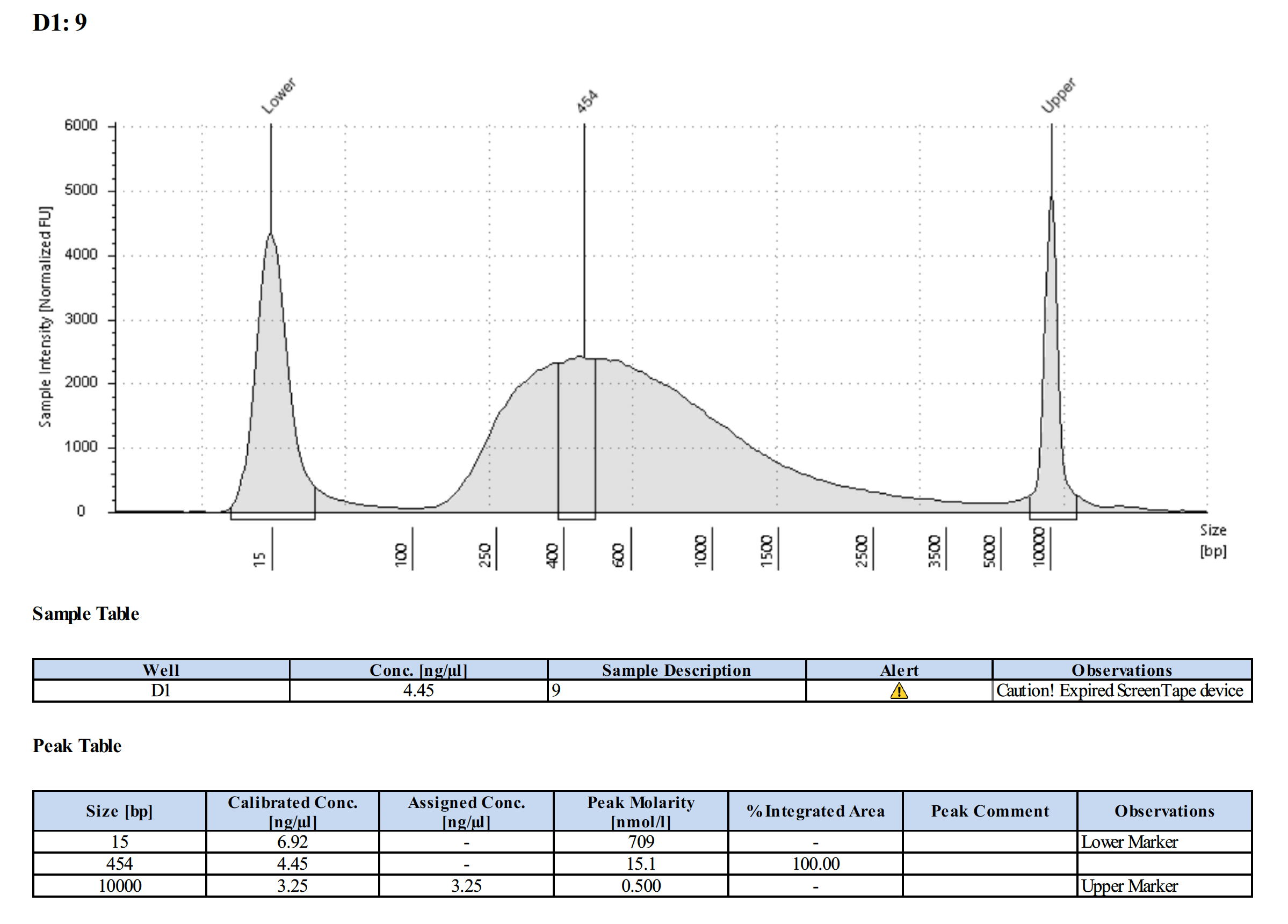
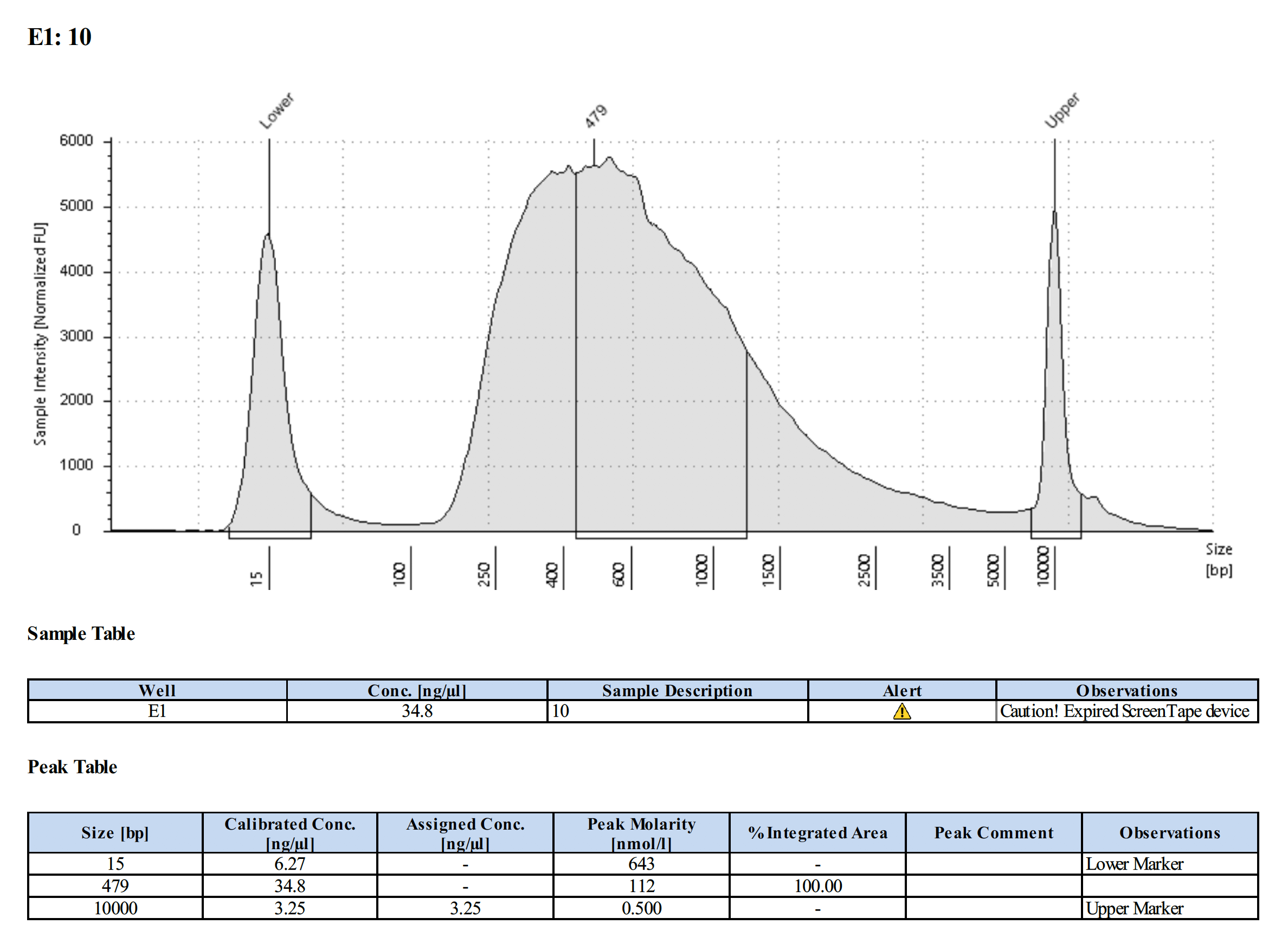
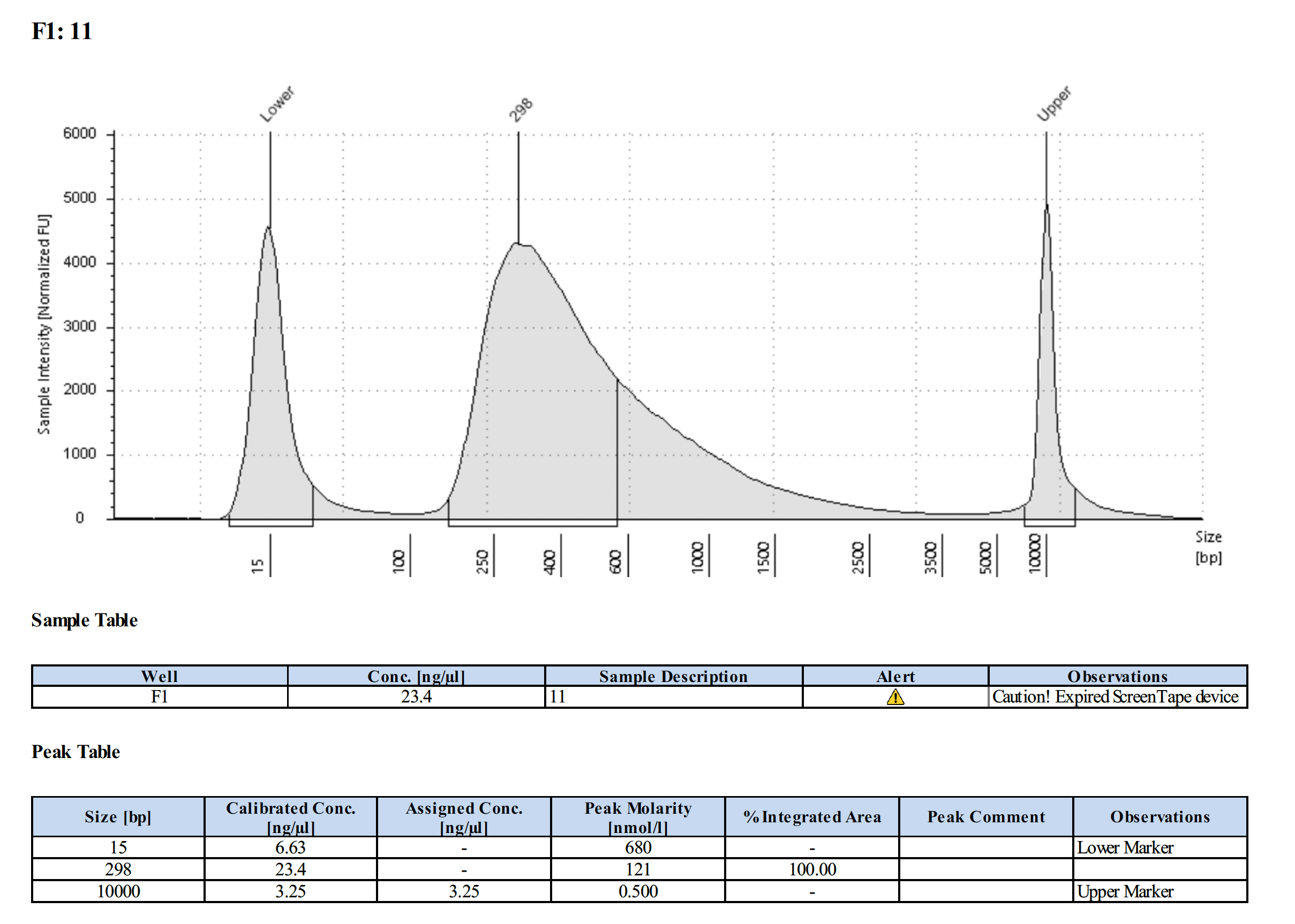
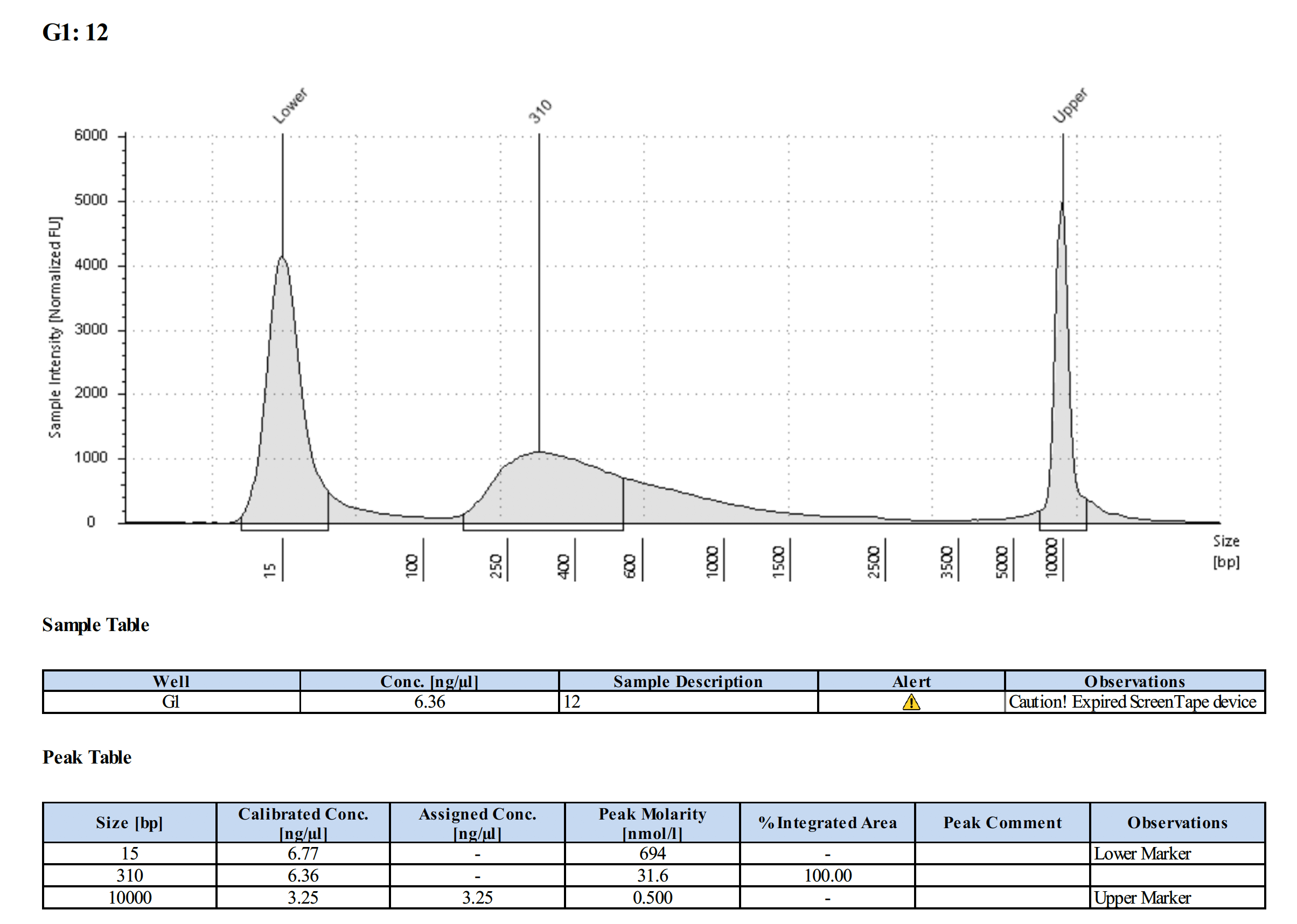
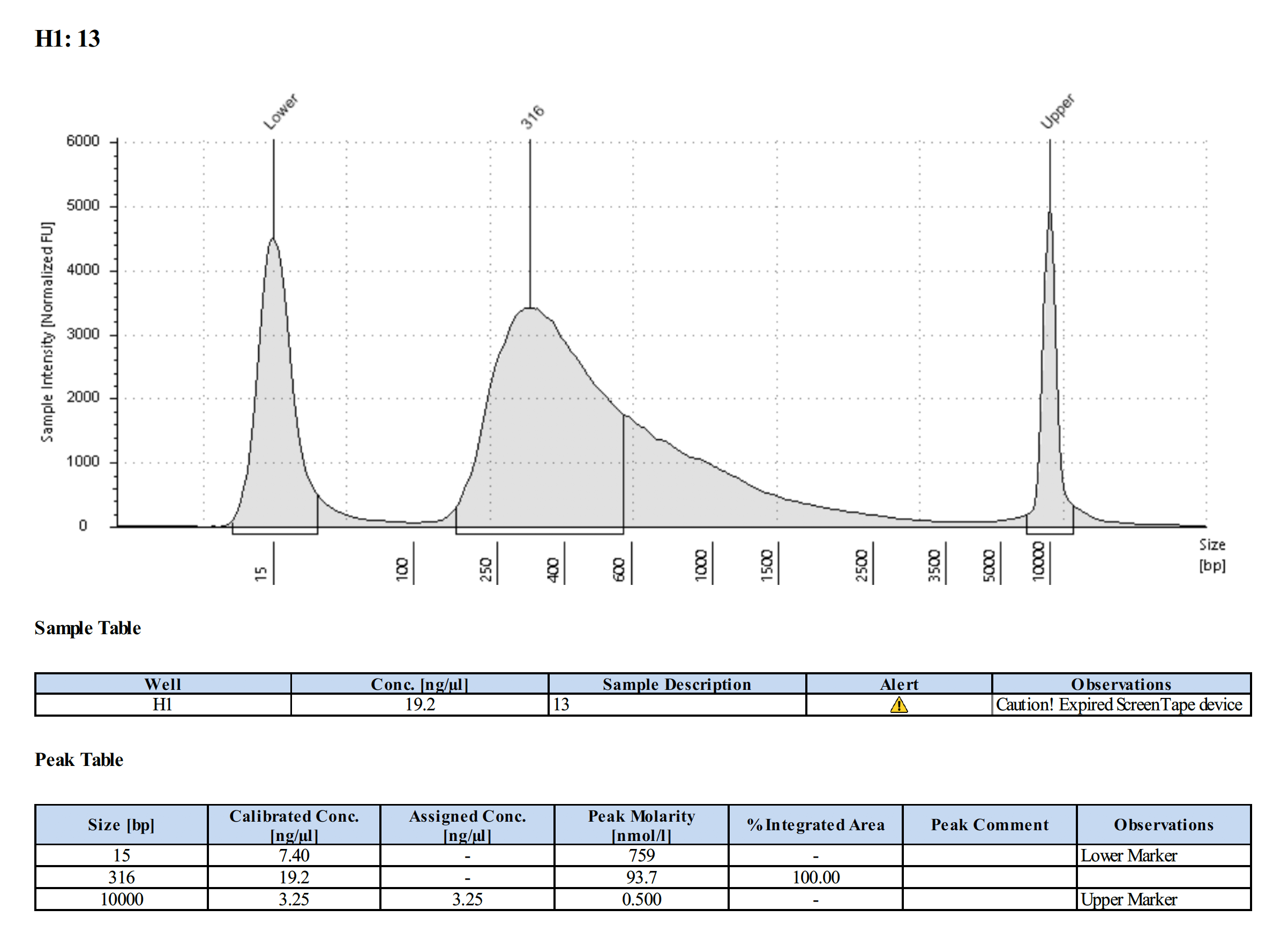
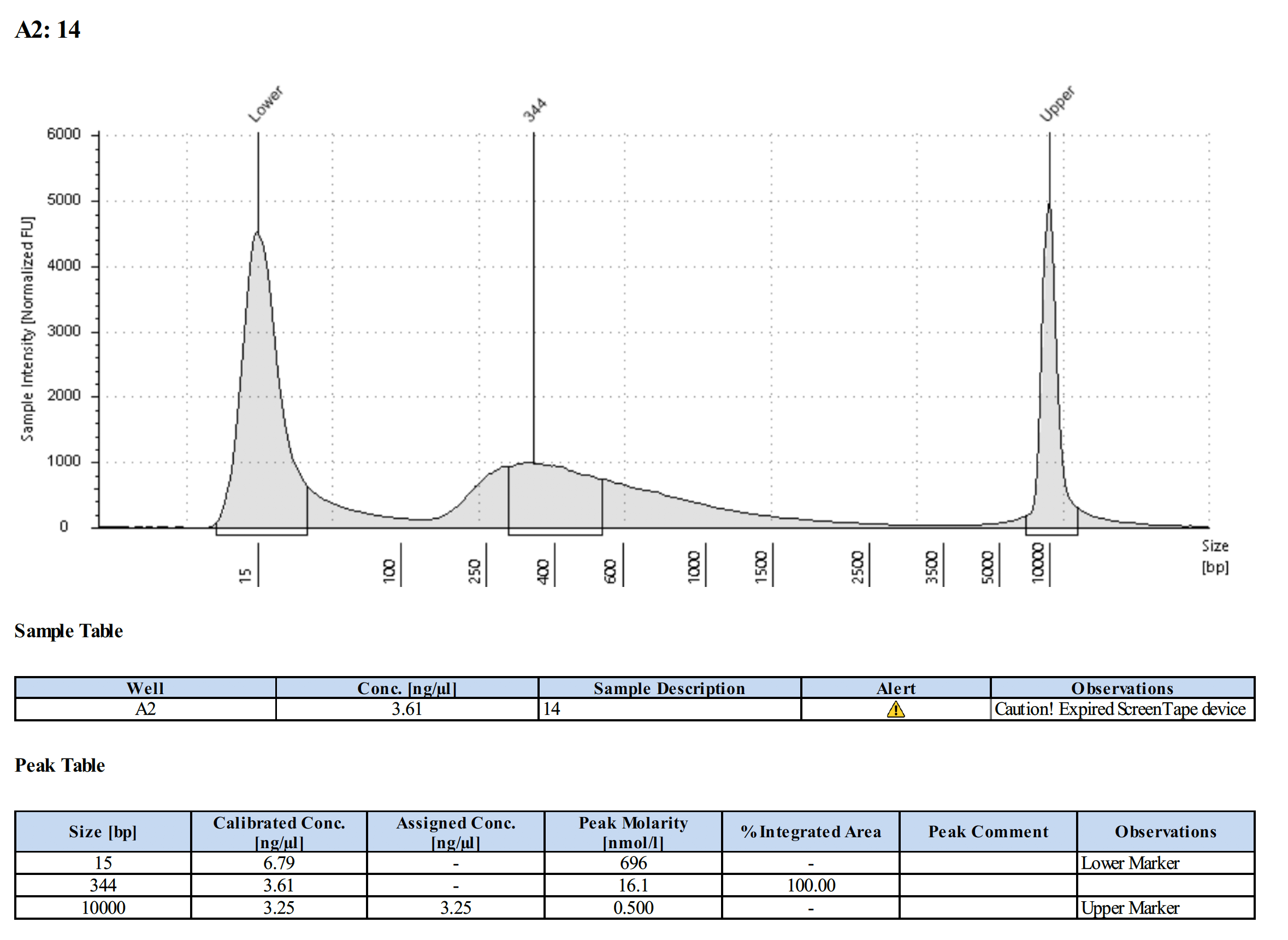
Summary
Both modifications produced good libraries. The KAPA HiFi modification increased the yield. This also occurred in previous library preps.
