20210510 WGBS PicoMethyl-seq library re-amplification
Project: Thermal Transplant Molecular
Goal
Re-amplify PicoMethyl-seq libraries that had low output (<5ng/ul) from initial prep using this protocol.
Protocol
Repeating the last two steps of this protocol:
1) Second amplification with 4 cycles instead of 10 cycles
2) 1X bead clean up
For all samples, I used the following inputs for the second amplification:
- 12 uL of the PicoMethyl-seq library
- 13 uL of Library Amp Master Mix (2x)
- 1 uL of the combined i5 and i7 index primers (I used the same indices as the original library prep)
Samples, indices, original Qubit quantification, post re-amplification Qubit quantification
| Sample # | i5/i7 Index # | Original Library (ng/uL) | Re-amp DNA #1 (ng/uL) | Re-amp DNA #2 (ng/uL) |
|---|---|---|---|---|
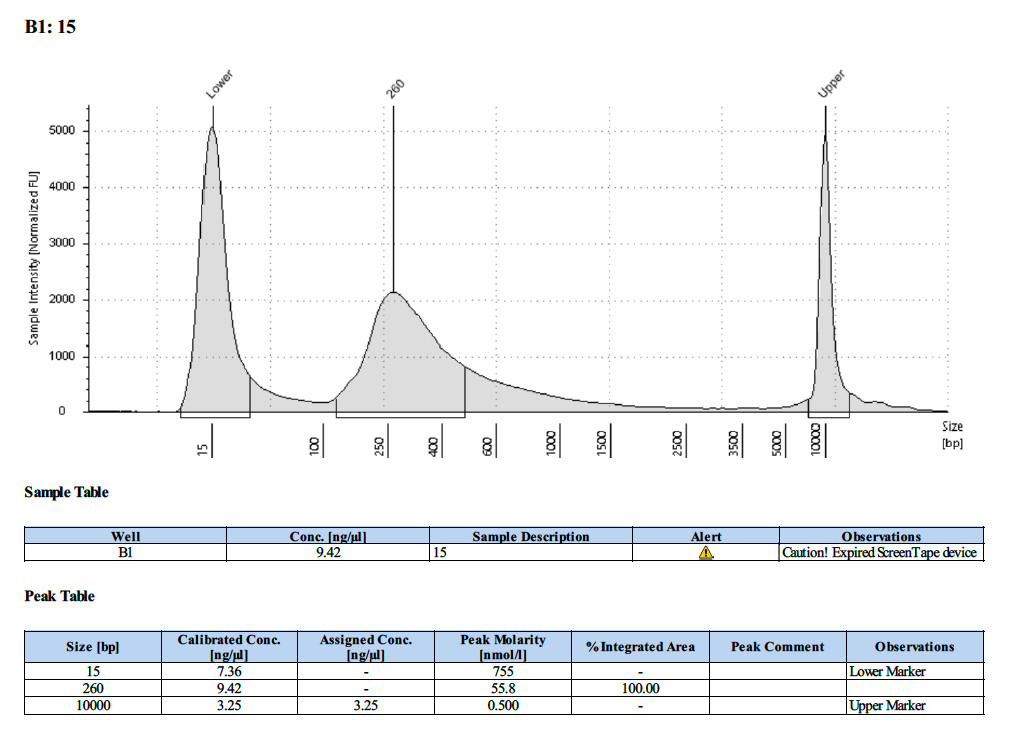
| 15 | 15 | LOW | 9.56 | 9.32 |
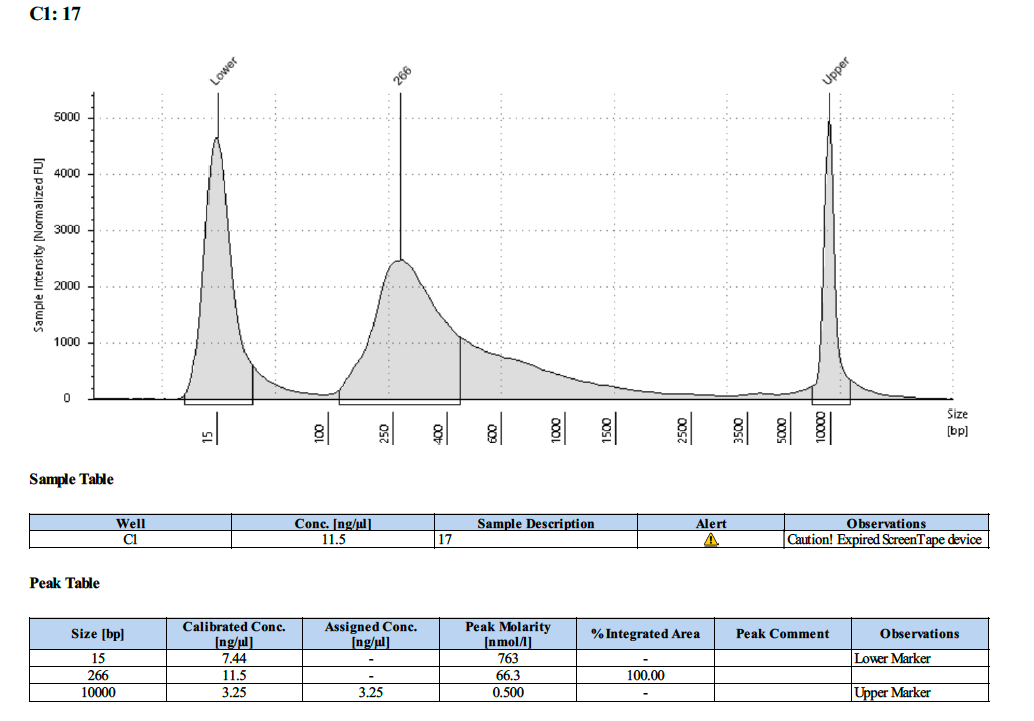
| 17 | 17 | 5.81 | 14.2 | 13.9 |
| 19 | 19 | LOW | LOW | LOW |
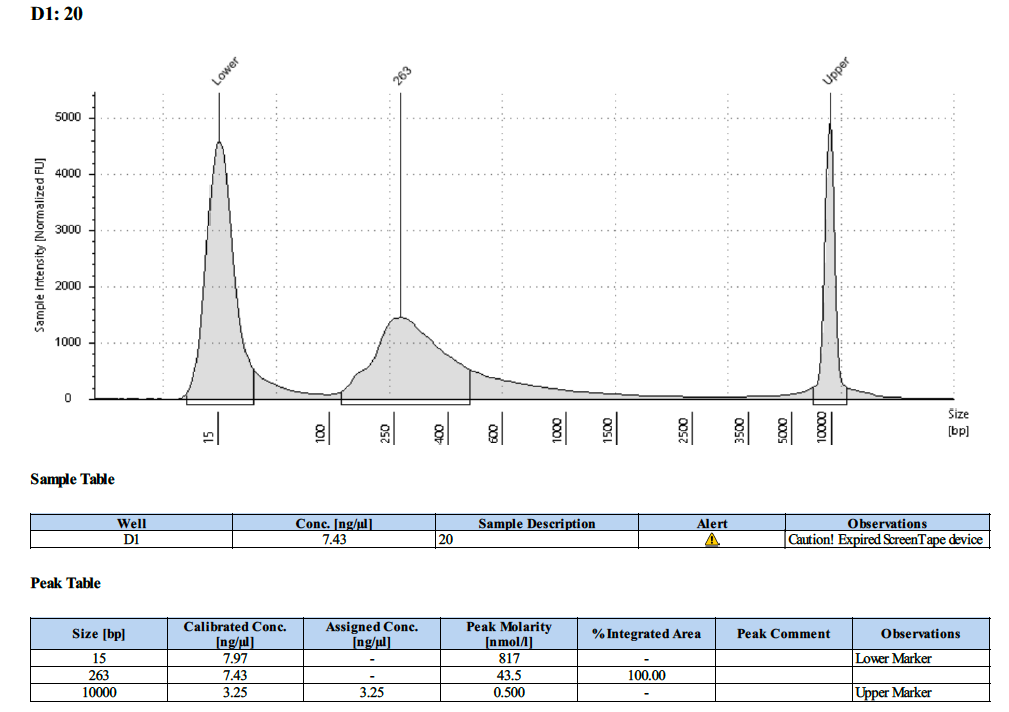
| 20 | 20 | 3.24 | 7.12 | 7.00 |
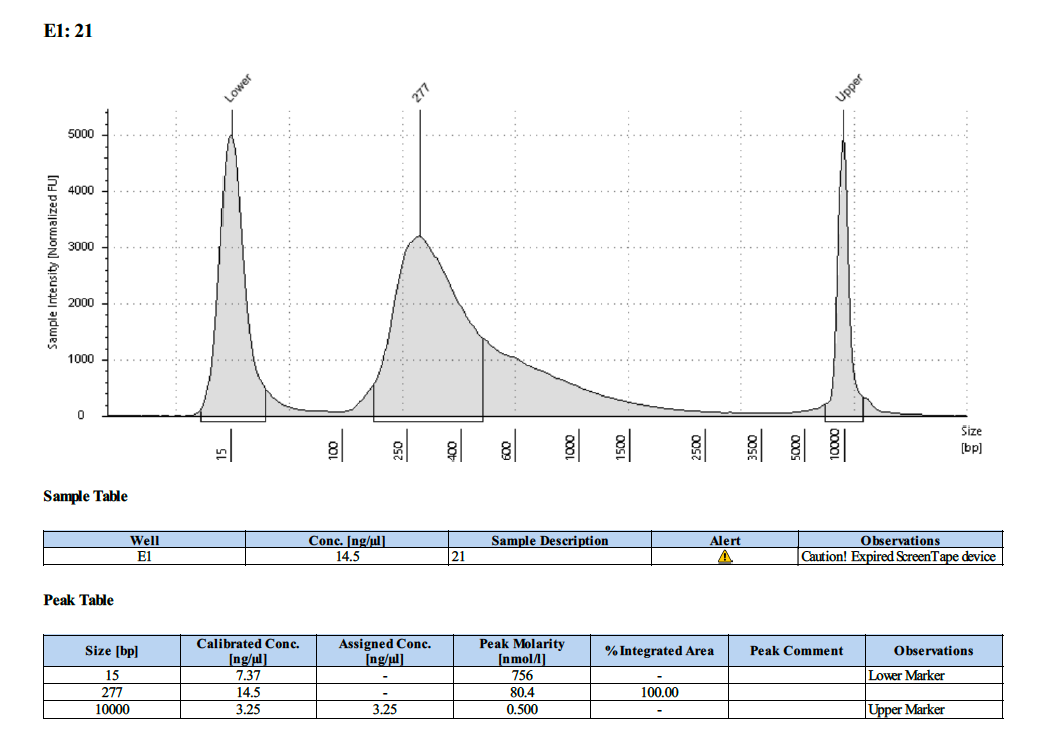
| 21 | 21 | 4.69 | 16.5 | 16.1 |
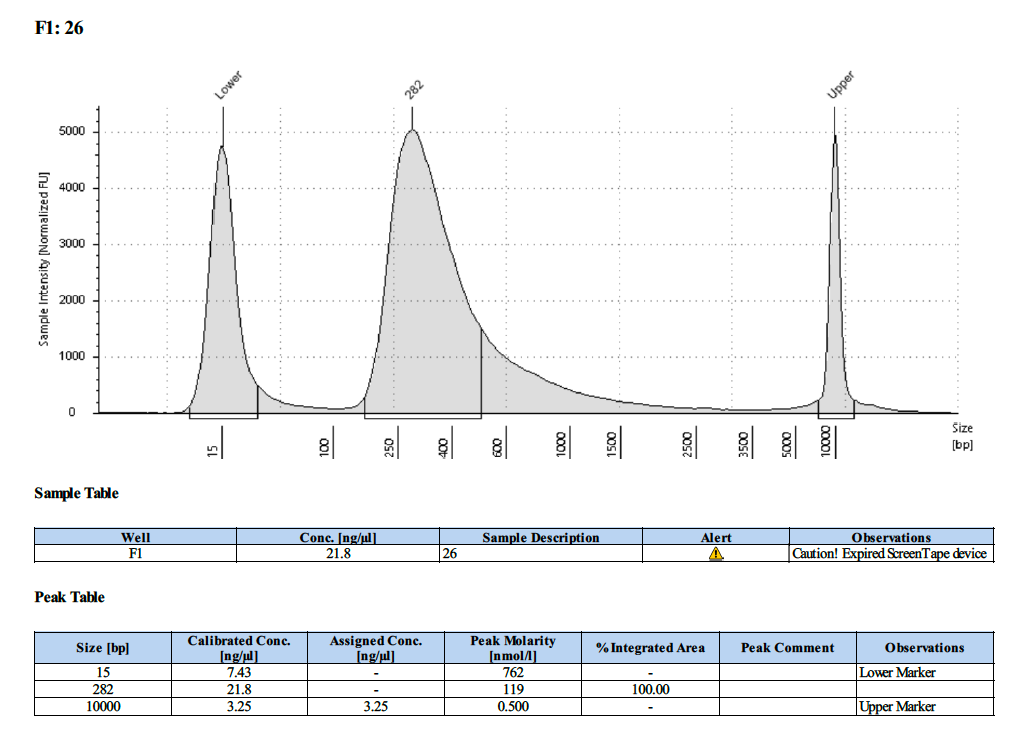
| 26 | 26 | 3.54 | 19.1 | 18.6 |
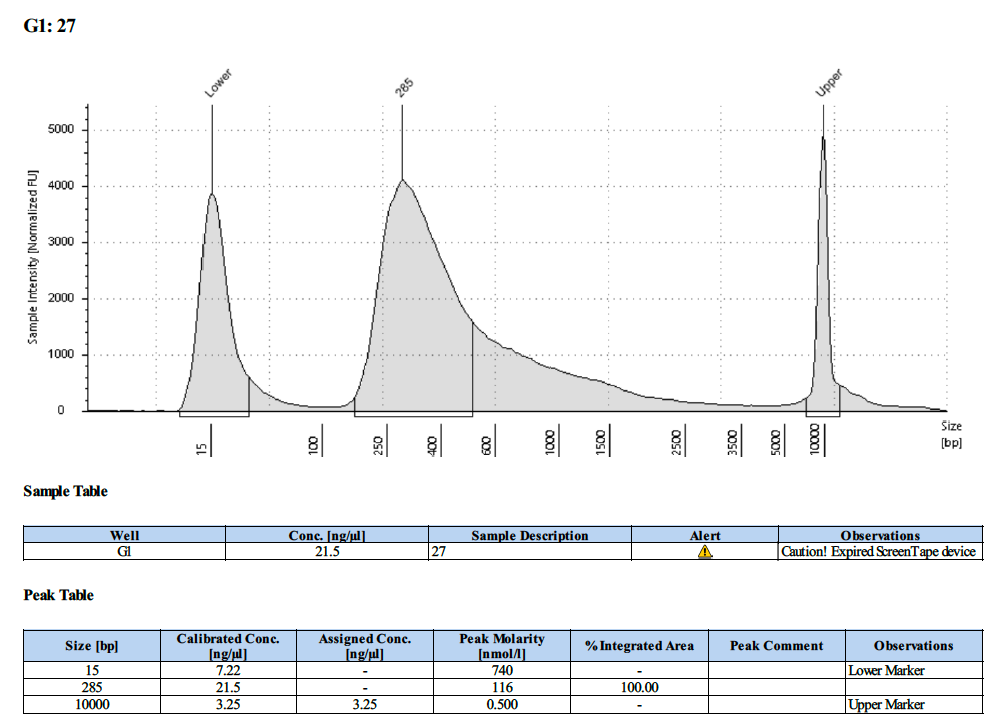
| 27 | 27 | 5.44 | 20.6 | 19.8 |
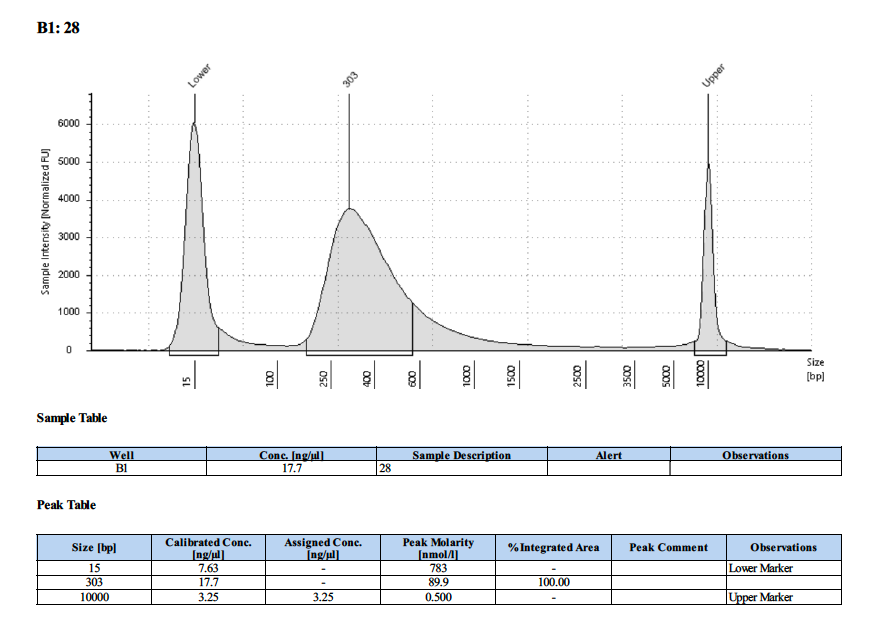
| 28 | 28 | 2.86 | 17.5 | 17.2 |
| 32 | 32 | 5.57 | LOW | LOW |
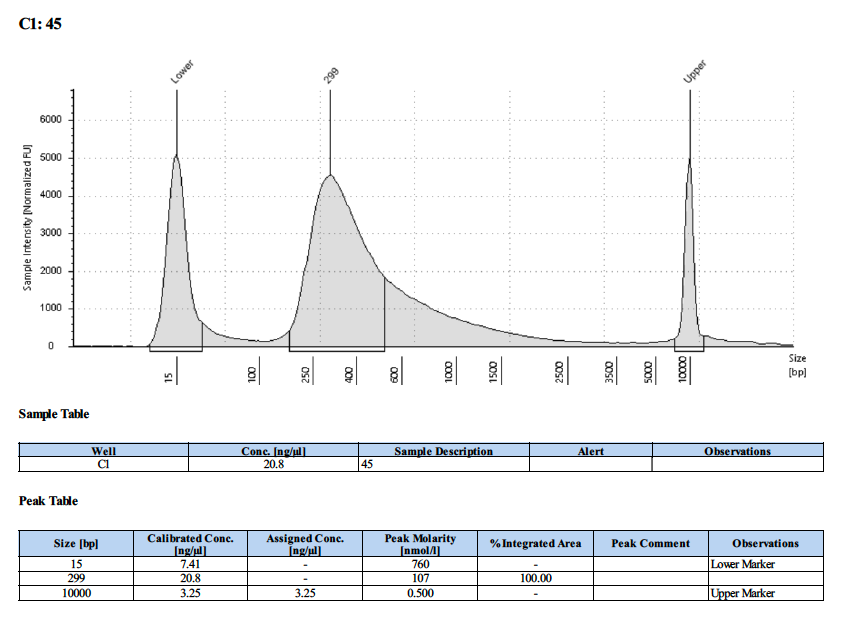
| 45 | 45 | 5.4 | 21.2 | 20.8 |
| Qubit std.1 | 178.09 | |||
| Qubit std.2 | 19930.40 |
Tapestation Results
Written on May 10, 2021
